
IMGT/GeneInfo Documentation
IMGT®, the international ImMunoGeneTics information system®
IMGT/GeneInfo provides information on data resulting from the mechanisms of V-J and V-D-J gene rearrangements in the T cell receptor (TR) loci of Homo sapiens and Mus musculus.
IMGT/GeneInfo query is made in 2 steps through 2 pages:
In the first screen, select the species (Homo sapiens or Mus musculus), the locus (TRA or TRB), and the gene combination (V-V, V-J or V-D-J).
You can either choose combinations corresponding to existing rearrangements, or "virtual" non-existing combinations given for informational purposes, (e.g. V-V combinations).
Once the desired combination is selected, click on the "submit" button.

Depending on your preceding choice, you can select the genes (V,D,J) for which information is queried, according to the gene name (official IMGT nomenclature or previous ones), or the relative position of the gene within the locus (number prefixing each gene name).
Once your selection is made, click on the submit button.

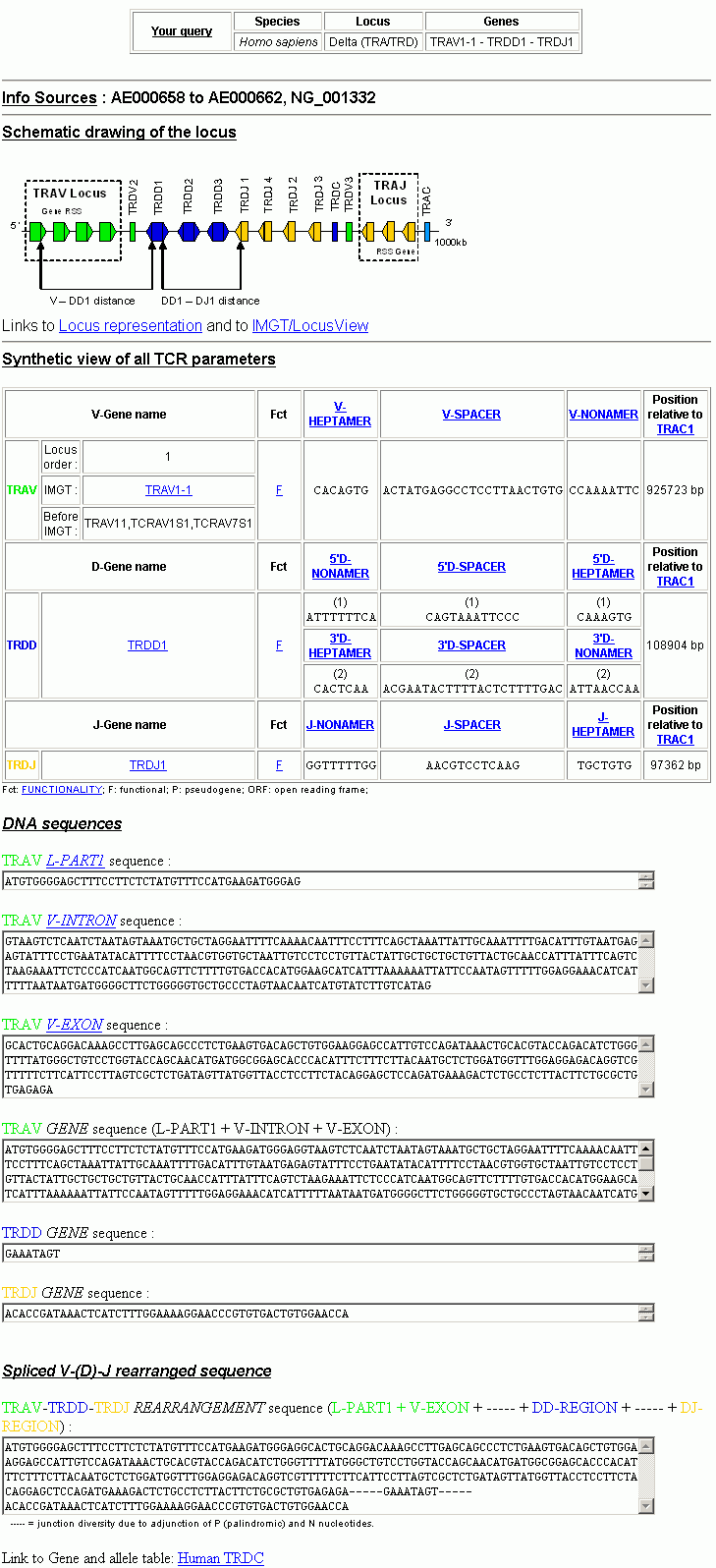
The IMGT/GeneInfo results page is divided in seven parts.
Part one (your query) is a reminder of what you asked for (species, locus, genes).
Part two (info sources) is the source from which information was collected (e.g. AE000658 for the human TRA/TRD locus).
Part three (schematic drawing of the locus) is an image that corresponds to the selected combination of genes and that explains visually which gene types are concerned, how the genes and the RS are oriented, and how distances between genes were computed.
Part four (synthethic view of all TCR parameters) is a table that contains, for each gene, a summary with the gene name, the functionality (functional, pseudogene, open reading frame) and the nucleotide sequences for each RS part (heptamer, spacer, nonamer). It also contains the corresponding consensus sequence when it exists; the position relative to the first base of: TRAC for the TRA/TRD loci and TRBC2 for the TRB loci; and the genomic distance in base pairs between the genes of the selected combination, in their germline configuration (from V-last base to J-first base).
Part five (DNA sequences) corresponds to the sequences of the gene and, for a V gene, to its various parts (L-PART1, V-INTRON, V-EXON), sequences can be selected for copy and paste. A color code is associated to all information originating from the same gene to make it easier to see and remember.
Part six (Spliced V-(D)-J rearranged sequence) is the complete rearranged and spliced sequence.
Part seven (Link) is a link towards additional and complementary information in the IMGT site.
For enquiries concerning IMGT/GeneInfo, please contact Thierry-Pascal BAUM
Software material and data concerning IMGT/GeneInfo may be used for academic research only, provided that it is referred to TIMC(1) and ICH(2), and cited as "IMGT/GeneInfo".
 |
(1) TIMC Laboratoire TIMC - IMAG - CNRS UMR 5525, Techniques de l'Imagerie, de la Modélisation et de la Cognition, Université Joseph Fourier, Grenoble 1 Faculté de Médecine, Domaine de la Merci, 38706 La Tronche, France. E-mail: tpbaum@imag.fr |
(2) ICH Laboratoire d'Immunochimie, CEA-G/DRDC/ICH CEA-Grenoble, INSERM U548 Université Joseph Fourier, Grenoble 1 17 rue des Martyrs, 38054 Grenoble Cedex 09, France. E-mail: IMMUNO@dsvsud.cea.fr |
 |
(3) LIGM Laboratoire d'ImmunoGénétique Moléculaire, LIGM, UPR CNRS 1142, IGH Université Montpellier 2, 141 rue de la Cardonille, 34396 Montpellier Cedex 5, France. E-mail: Marie-Paule.Lefranc@igh.cnrs.fr |