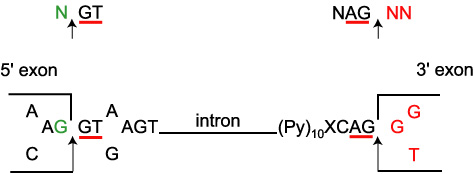
Splicing sites
| IMGT labels: | DONOR-SPLICE | ACCEPTOR-SPLICE |
|---|---|---|
| Colored letters in that figure correspond to splicing frame 1 |
|
|
Introns always have two distinct nucleotides at either end. At the 5' end the DNA nucleotides are GT [GU in the premessenger RNA (pre-mRNA)]; at the 3' end they are AG. These nucleotides are part of the splicing sites.
DONOR-SPLICE: splicing site at the beginning of an intron, intron 5' left end.
ACCEPTOR-SPLICE: splicing site at the end of an intron, intron 3' right end.
The GT/AG mRNA processing rule is applicable for almost all eukaryotic genes [1,2].
A polypyrimidine (CnTn) motif is present upstream of the CAG intron 3' ending. More upstream, the consensus branch site (CTGAC) (not shown) is a necessary component in the effective splicing of the pre-mRNA [3].
The snRP's or "snurps" (small nuclear ribonucleoproteins) search out the sequences above and join together with other snRP's to form a spliceosome. These cut out the introns, forming the "lariat formation" of the excised intron.
Once introns are removed , the mature messenger RNA (mRNA) leaves the nucleus and is translated into protein (Protein synthesis).
| Splicing INTRON view | Splicing EXON view | ||||
|---|---|---|---|---|---|
| Different splicing types | In IMGT/LIGM-DB flat file for exon translation(1) |
In IMGT/GENE-DB reference sequences of exons in FASTA format |
|||
| Number of 'd' nucleotides (5) | |||||
| added in 5' of the exon | deleted in 3' of the exon | ||||
| Usual splicing for IG and TR (except M exons) (2) |
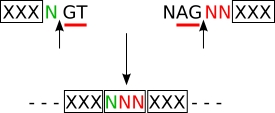 |
splicing frame 1 (sf1) |
codon_start3 (translation frame 3) |
1 | 1 |
| Splicing for M exons (3) |
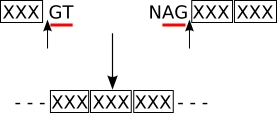 |
splicing frame 0 (sf0) |
codon_start1 (translation frame 1) |
0 | 0 |
| Other splicing (4) |
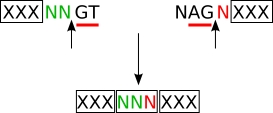 |
splicing frame 2 (sf2) |
codon_start2 (translation frame 2) |
2 | 2 |
Arrows mark exon-intron (DONOR-SPLICE) and intron-exon (ACCEPTOR-SPLICE) boundary.
X represent nucleotides of a complete exon.
N represent nucleotides that make up the codon resulting from the splicing.
The nucleotide(s) N of a donor exon (in green in the table) is(are) designated as 'd' nucleotides.
Splicing INTRON view
The DONOR-SPLICE and the ACCEPTOR-SPLICE of an intron (between two exons to be spliced)
must belong to the same splicing frame.
For examples:
- the splicing frames of the splicing sites of a V-INTRON are both of type 1, and the V-INTRON is defined as in splicing frame 1,
- the splicing frames of the splicing sites at the beginning and end of the intron between M1 and M2 exons of IGHC genes are both of type 0, and this intron is defined as in splicing frame 0.
If the two splicing sites of an intron are not in the same frame, the splicing will disrupt the coding reading frame in the second downstream (3') exon and will create a frameshift.
Splicing EXON view
There is no requirement for the splicing sites at the ends of an exon to match. For example, an exon can have an
ACCEPTOR-SPLICE in splicing frame 1 (5'nt-sf1), and a DONOR-SPLICE in splicing frame 0 (3'nt-sf0).
If the splice sites at the ends of an exon happen to match, that exon will have a number of base pairs that is an integer multiple of 3.
If those splice sites happen not to match, the exon will not have a number of base pairs that is an integer multiple of 3.
For that reason and to bridge the gap between nucleotide and amino acid sequences in the IMGT/GENE-DB reference sequences of exons, in FASTA format, the 5' end of an exon is completed with the 'd' nucleotides of the upstream donor exon and its 3' end is deleted from its own 'd' nucleotides.
Representations in spliced sequences
- splicing frame 1
E (G) Q gaa g/gt cag
- splicing frame 2
K (S) I aag ag/c atc
- splicing frame 0
M/N P atg/aat ccc
Legend:
Small vertical arrows or lines can indicate the limits of the spliced exons.
Colored letters can be used (IMGT Color menu for splicing types).
Enventually underlined letters can indicate the codon resulting from the splicing.
A slash can indicate the limit of the exons.
Amino acids resulting from the splicing can be shown between parentheses.
Exemples are from TREML1.
| (1) | Codon_start refers to the position of the first nucleotide of the first complete codon in the genomic 3' exon. This information is used for the IMGT/LIGM-DB automatic in-frame translation tool. |
| (2) | The codon NNN which results from the splicing is encoded by the last nucleotide (designated as 'd') of the donor exon and the first two nucleotides of the acceptor exon. |
| (3) | There is no new codon resulting from the splicing. |
| (4) | The codon NNN which results from the splicing is encoded by the two last nucleotides (designated as 'd') of the donor exon and the first nucleotide of the acceptor exon. |
| (5) | Number of nucleotides added at the 5' end of the exon, and/or deleted at the 3' end of the exon, to obtain complete codons at both ends. These numbers depend on the respective splicing frames. |
| [1] | Shapiro, M.B. and Senapathy, P., Nucleic Acids Res., 15:7155-7174 (1987) PMID: 3658675 |
| [2] | Burset, M. et al., Nucleic Acids Res., 29:255-259 (2001) PMID 11125105 |
| [3] | Maniatis, T. and Reed, R., Nature, 325:673-678 (1987) PMID: 2950324 |
- IMGT Color menu for splicing types (IMGT Scientific chart)



