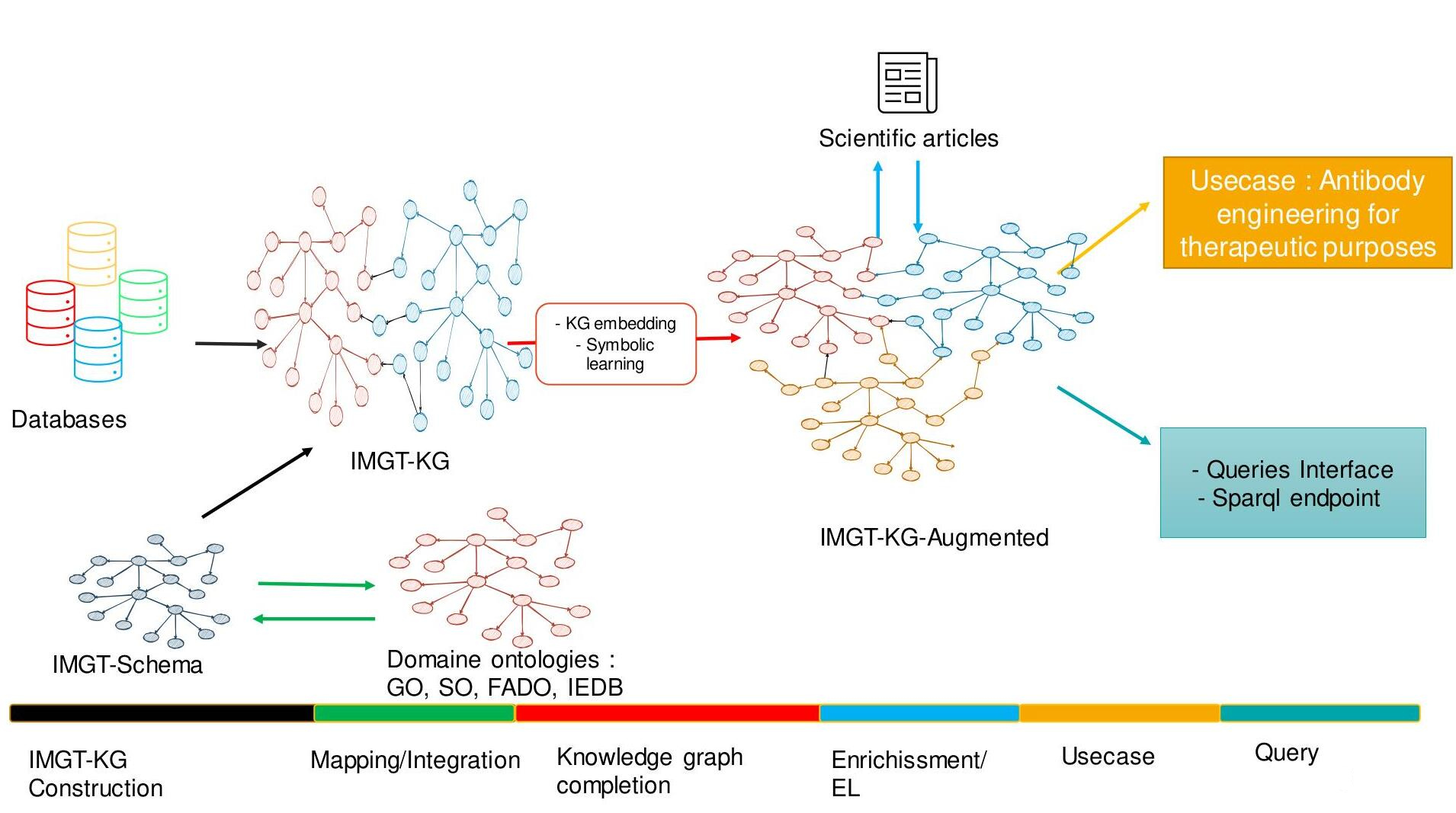
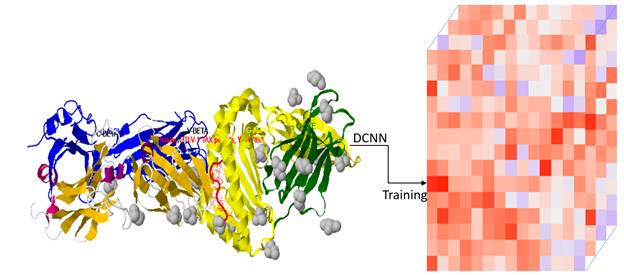
Recognition and prediction of the immunoglobulins and T cell receptors at the genomic level
The Human Genome Project, which remains the world's largest collaborative genetics project was launched in 1990
and it was declared complete in 2003. Since then, the methodological and technological advances in the field
have made it possible to obtain the whole genome sequencing of a species in much shorter time and in much
deeper detail compared to the first version of the human genome. Nowadays, several species genomes are fully
sequenced and accessible via the publicly available databases. The adaptive immune response, which appeared
450 million years ago, is found in all jawed vertebrates, that is in species going from fish to human.
The identification and the annotation of the immunoglobulins (IG) and T cell receptors (TR) which are notorious
for high similarity among themselves constitutes a tedious and time-consuming process which is also prone to
mistakes. The current state of the art is relying on sequence alignments for the successful identification
and annotation of the IG and TR.
In the under question project and in collaboration with ATOS, Deep Learning
algorithmic approaches are explored to see if the identification and annotation of the immunoglobulins
and T cell receptor genes can be achieved with more efficiency in comparison to the traditional sequence
alignment based methodologies. Traditional and new Deep Learning architectures are being used such as
Deep Neural Network (DNN), Convolutional Neural Network (CNN), Recurrent Neural Network (RNN), Attention
mechanism, Transformers and BERT to address the immunogenetic questions. The complete set of IG and TR
of a genome apart from addressing fundamental questions in the fields of molecular evolution and
comparative genomics it provides the necessary ground information to enter, for instance, into the
personalized medicine as well as the livestock enhancement fields.