IMGT/StatClonotype Documentation
https://www.imgt.org
IMGT®, the international ImMunoGeneTics information system®

| Introduction |
 IMGT/StatClonotype works only with uploaded files (.txt format) from the IMGT/HighV-QUEST statistical analysis output ('stats_xxx' file available in 'data' directory of the IMGT/HighV-QUEST statistical analysis output where 'xxx' is the batch name and the locus type) as described in section B.
IMGT/StatClonotype works only with uploaded files (.txt format) from the IMGT/HighV-QUEST statistical analysis output ('stats_xxx' file available in 'data' directory of the IMGT/HighV-QUEST statistical analysis output where 'xxx' is the batch name and the locus type) as described in section B.| 'IMGTStatClonotype' R package installation |
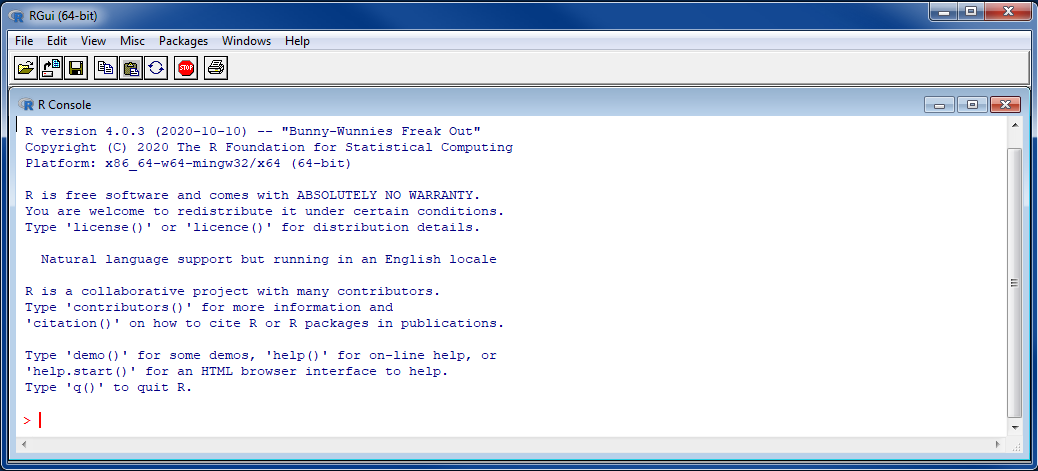
 Note that: If R is already installed on your computer please check if you have a recent version (>=4.0.3).
Note that: If R is already installed on your computer please check if you have a recent version (>=4.0.3).
 For Windows operating system
For Windows operating system
 install.packages(c("reshape2", "data.table", "ggplot2", "shiny", "shinyjs",
"colourpicker", "gridExtra", "DT", "plotly", "htmlwidgets", "png", "dendextend"))
# Choose the website you want to download the package from, you should choose "France" (or another country)
install.packages(c("reshape2", "data.table", "ggplot2", "shiny", "shinyjs",
"colourpicker", "gridExtra", "DT", "plotly", "htmlwidgets", "png", "dendextend"))
# Choose the website you want to download the package from, you should choose "France" (or another country) wspace <- getwd()# Press ENTER
wspace <- getwd()# Press ENTER  dest =paste(wspace,"/d3heatmap_0.6.1.tar.gz",sep="")# Press ENTER
dest =paste(wspace,"/d3heatmap_0.6.1.tar.gz",sep="")# Press ENTER  download.file("https://www.imgt.org/download/StatClonotype/d3heatmap_0.6.1.tar.gz",destfile=dest)# Press ENTER
download.file("https://www.imgt.org/download/StatClonotype/d3heatmap_0.6.1.tar.gz",destfile=dest)# Press ENTER  install.packages("d3heatmap_0.6.1.tar.gz", repos=NULL)# Press ENTER
install.packages("d3heatmap_0.6.1.tar.gz", repos=NULL)# Press ENTER  if (!requireNamespace("BiocManager", quietly = TRUE)) install.packages("BiocManager")# Press ENTER
if (!requireNamespace("BiocManager", quietly = TRUE)) install.packages("BiocManager")# Press ENTER  BiocManager::install("multtest") # If a message appears to update a list of package (Update all/some/none? [a/s/n]) type n then press ENTER
BiocManager::install("multtest") # If a message appears to update a list of package (Update all/some/none? [a/s/n]) type n then press ENTER des=paste(wspace,"/IMGTStatClonotype_1.0.4.tar.gz",sep="")# Press ENTER
des=paste(wspace,"/IMGTStatClonotype_1.0.4.tar.gz",sep="")# Press ENTER download.file("https://www.imgt.org/download/StatClonotype/IMGTStatClonotype_1.0.4.tar.gz",destfile=des)# Press ENTER
download.file("https://www.imgt.org/download/StatClonotype/IMGTStatClonotype_1.0.4.tar.gz",destfile=des)# Press ENTER  install.packages("IMGTStatClonotype_1.0.4.tar.gz", repos=NULL)# Press ENTER
install.packages("IMGTStatClonotype_1.0.4.tar.gz", repos=NULL)# Press ENTER 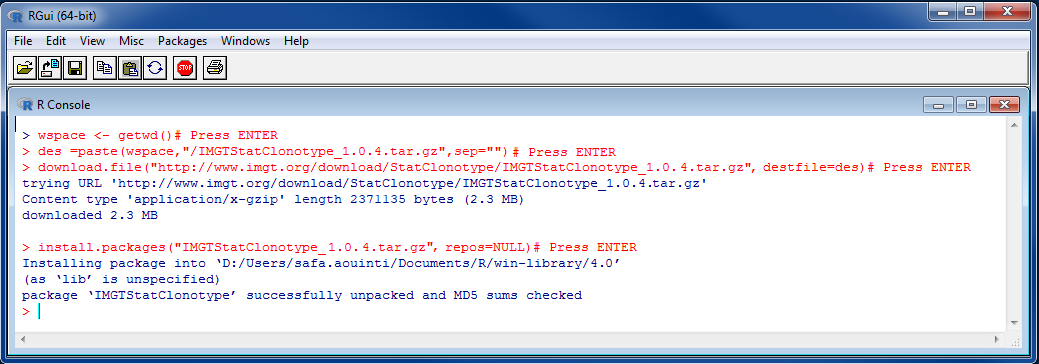
 See the user manual for more details: IMGTStatClonotype-manual
See the user manual for more details: IMGTStatClonotype-manual  For non-Windows operating system (Linux or Macintosh)
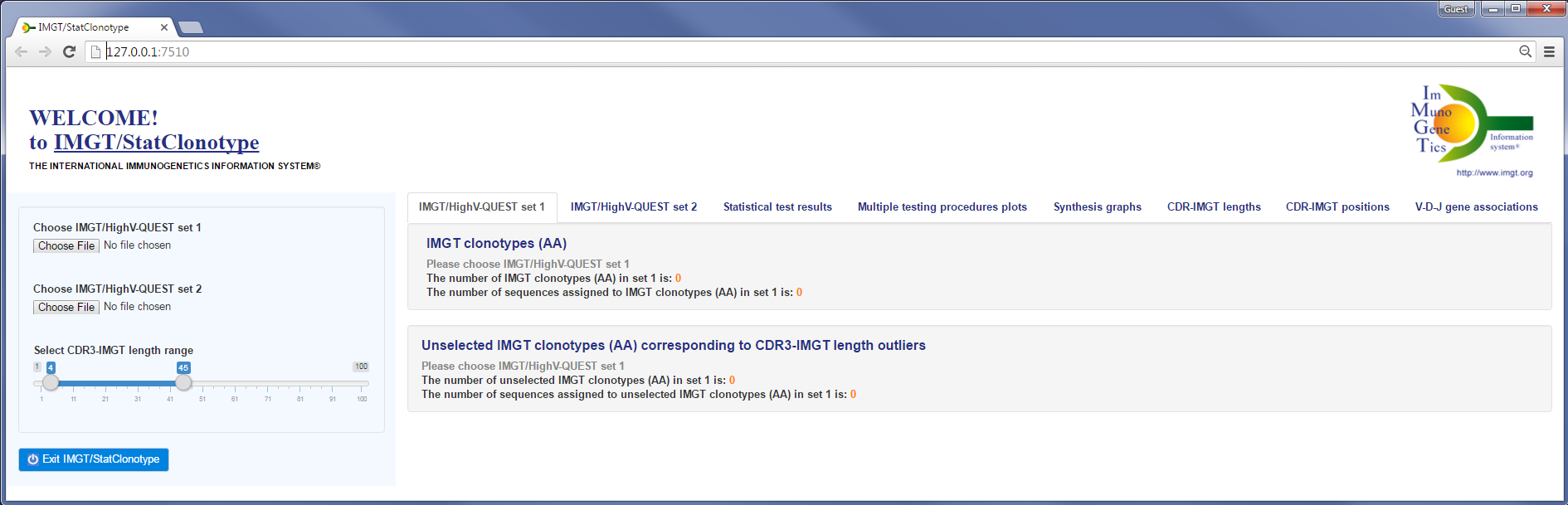
For non-Windows operating system (Linux or Macintosh)| IMGT/StatClonotype web tool |
 library(IMGTStatClonotype)# Press ENTER
library(IMGTStatClonotype)# Press ENTER  launch() # Press ENTER
launch() # Press ENTER
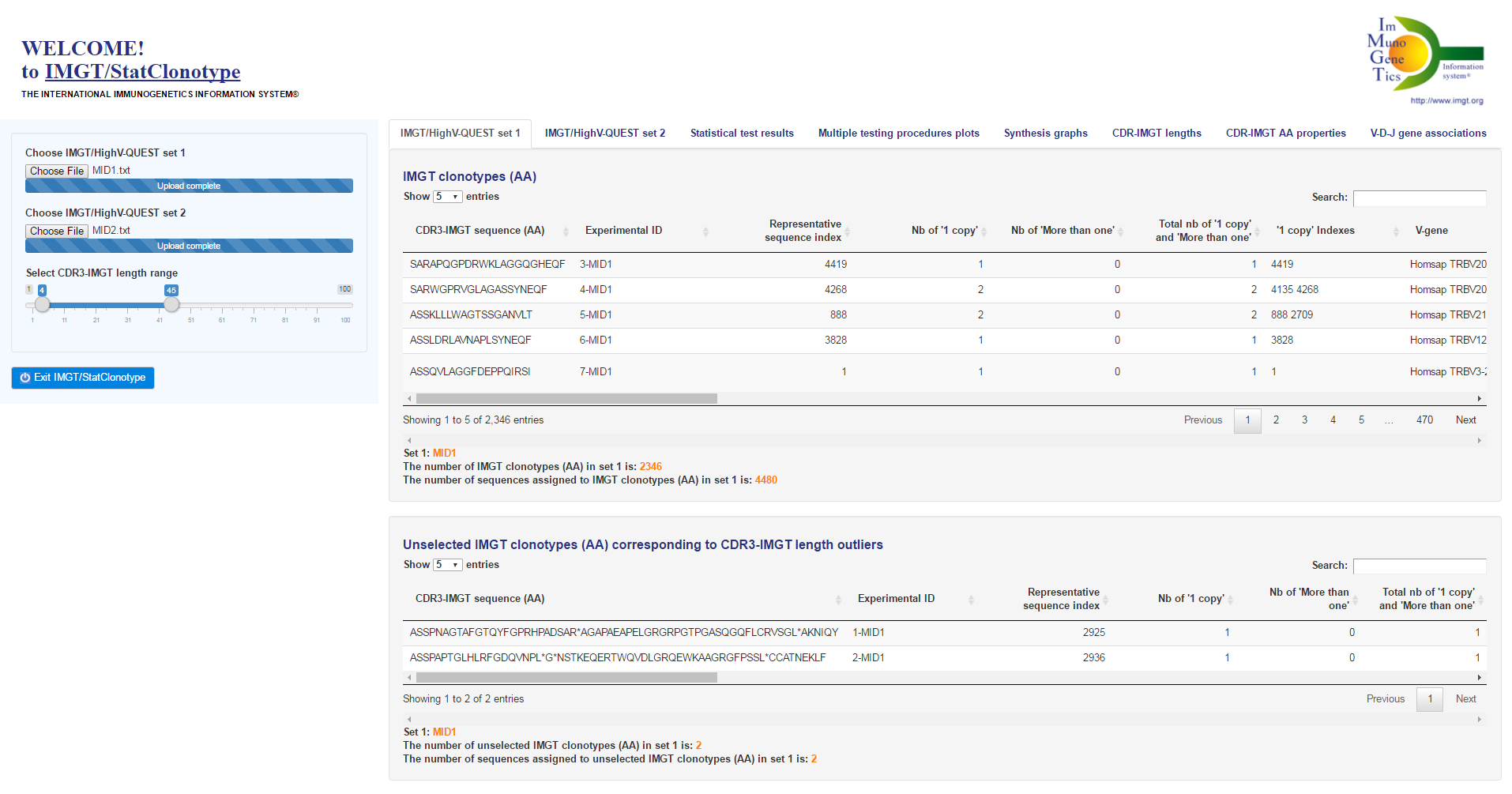
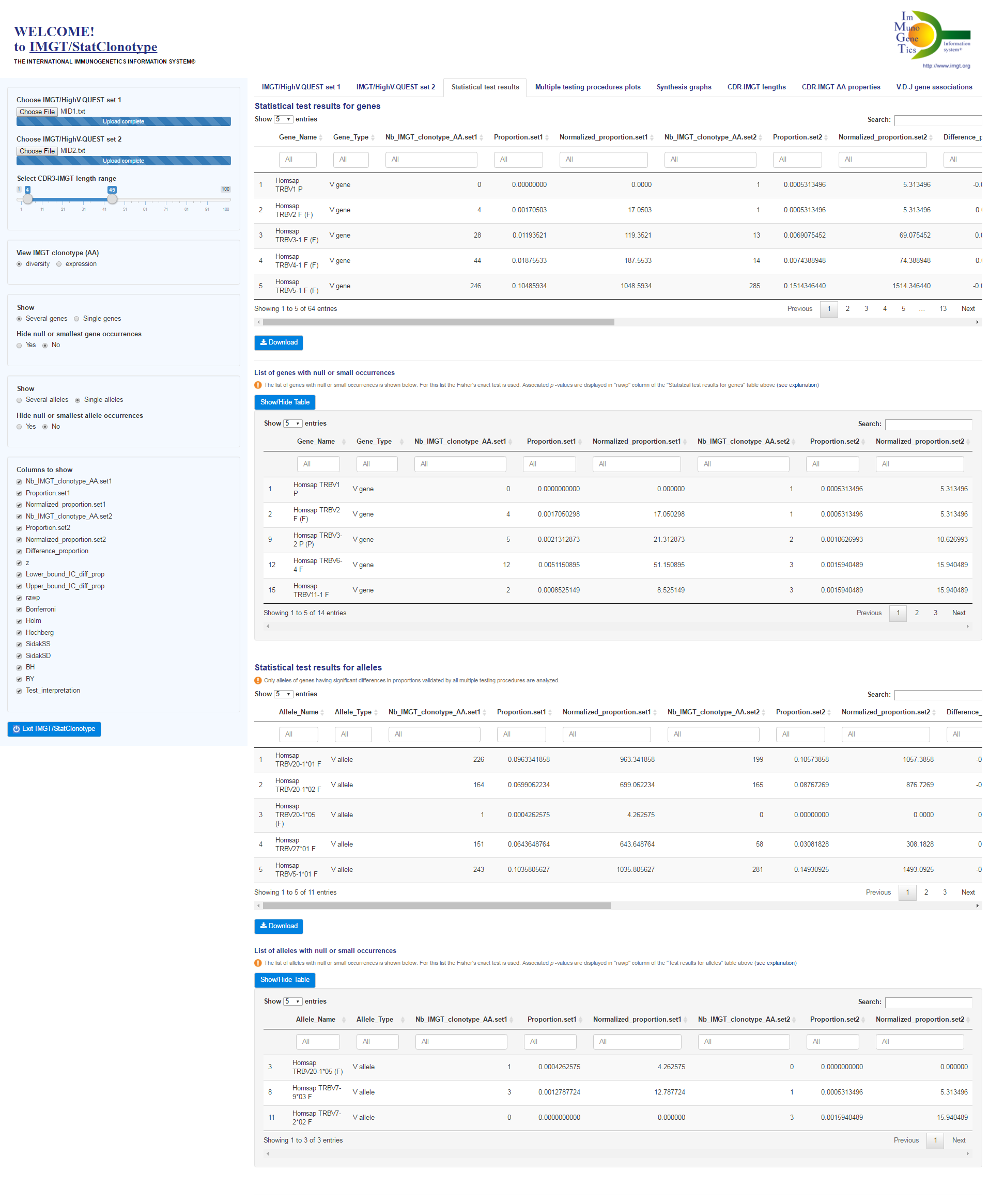
 Reminder: Properties of the multiple testing procedures (see [1] for more details)
Reminder: Properties of the multiple testing procedures (see [1] for more details)
| Procedures | Type of control | Algorithm structure | dependence of p-values under H0 | Properties |
| Bonferroni | FWER | Single-step | Ignorance | The most conservative |
| Šidák (SS) | FWER | Single-step | Independence | Less conservative than Bonferroni |
| Holm | FWER | Step-down | Ignorance | Less conservative than Bonferroni |
| Šidák (SD) | FWER | Step-down | Dependence | Similar to Holm |
| Hochberg | FWER | Step-up | Independence | Step-up of Holm |
| Benjamini & Hochberg (BH) | FDR | Step-up | Independence | The least conservative |
| Benjamini & Yekutieli (BY) | FDR | Step-up | Ignorance | More conservative than BH |
 We assume that the two compared sets are independent and the individual tests are independent of each other (i.e., multiple hypotheses are independent).
We assume that the two compared sets are independent and the individual tests are independent of each other (i.e., multiple hypotheses are independent).
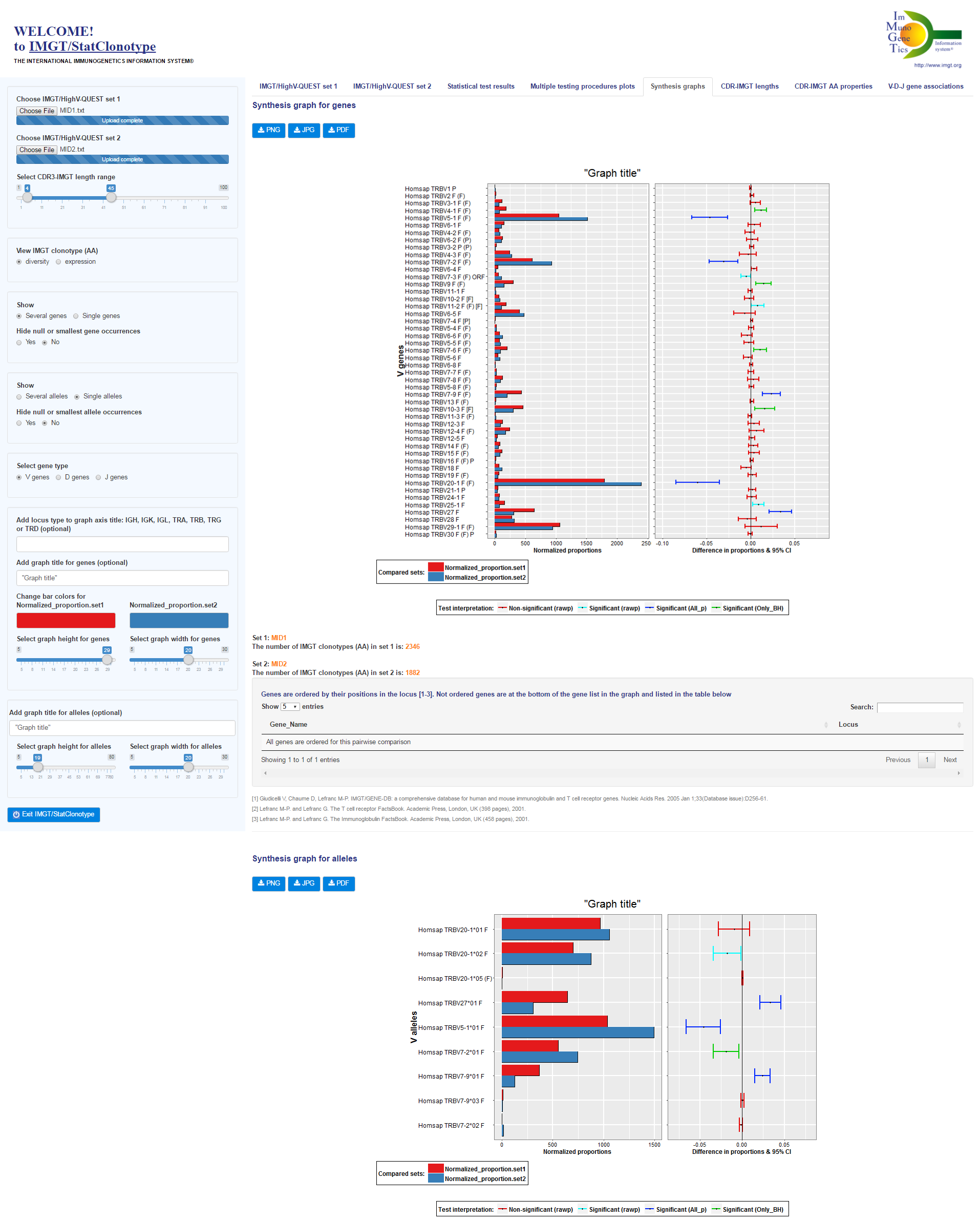
 For this list of genes the Fisher's exact test is used and associated p-values are displayed in "rawp" column of the "Test results for genes" table.
For this list of genes the Fisher's exact test is used and associated p-values are displayed in "rawp" column of the "Test results for genes" table.
 Only alleles of genes having significant differences in proportions validated by all multiple testing procedures are analyzed.
Only alleles of genes having significant differences in proportions validated by all multiple testing procedures are analyzed.
 For this list of alleles the Fisher's exact test is used and associated p-values are displayed in "rawp" column of the "Test results for alleles" table.
For this list of alleles the Fisher's exact test is used and associated p-values are displayed in "rawp" column of the "Test results for alleles" table.
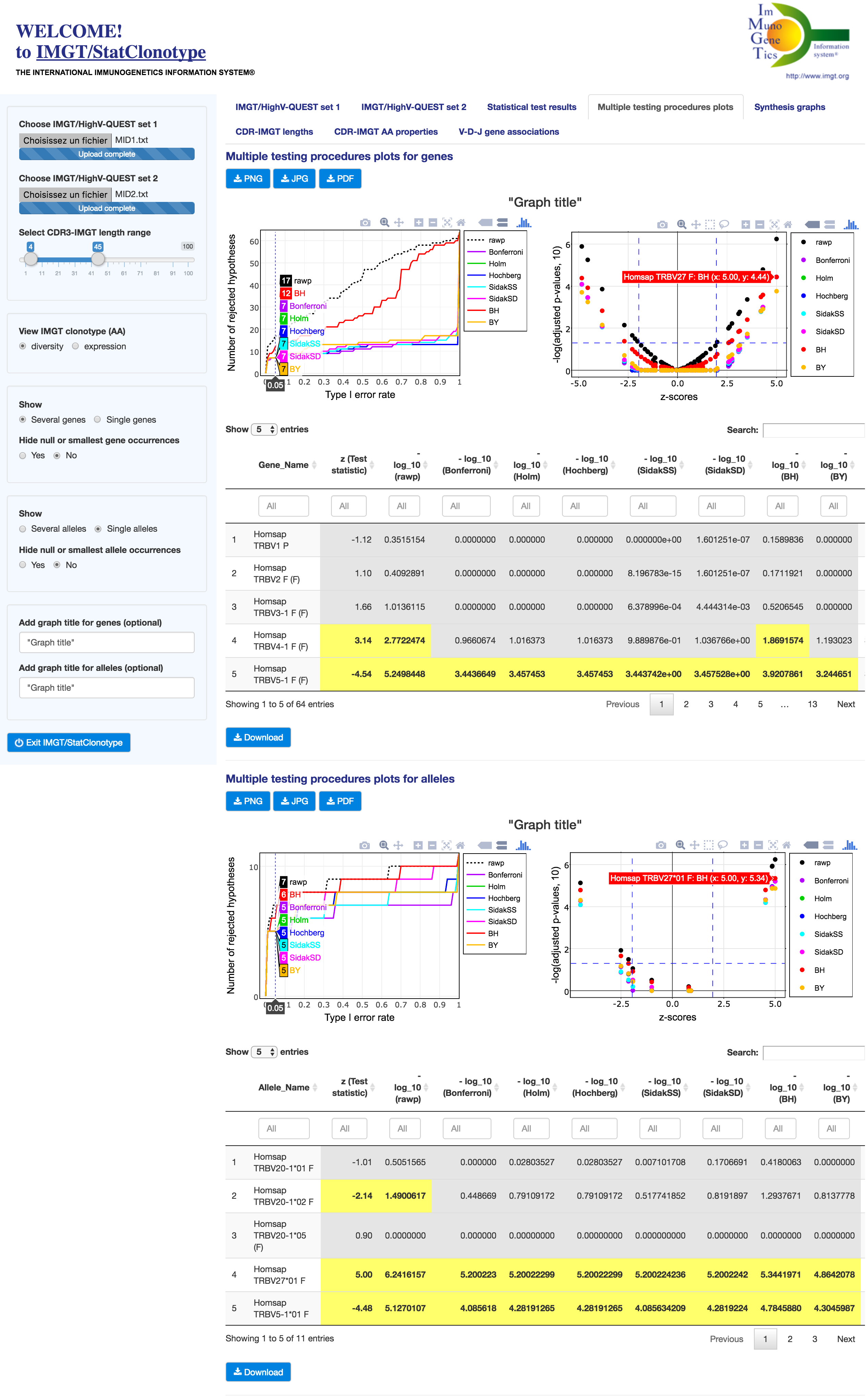
 Reminder: Only alleles of genes having significant differences in proportions validated by all multiple testing procedures are analyzed and visualized.
Reminder: Only alleles of genes having significant differences in proportions validated by all multiple testing procedures are analyzed and visualized.
 Note that:
Note that:
 Reminder: Only alleles of genes having significant differences in proportions validated by all multiple testing procedures are analyzed and visualized.
Reminder: Only alleles of genes having significant differences in proportions validated by all multiple testing procedures are analyzed and visualized.
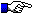 Note that: CDR-IMGT lengths (x-axis values) are not necessarily successive in graphs because only lengths found in one or both of compared sets are displayed.
Note that: CDR-IMGT lengths (x-axis values) are not necessarily successive in graphs because only lengths found in one or both of compared sets are displayed.
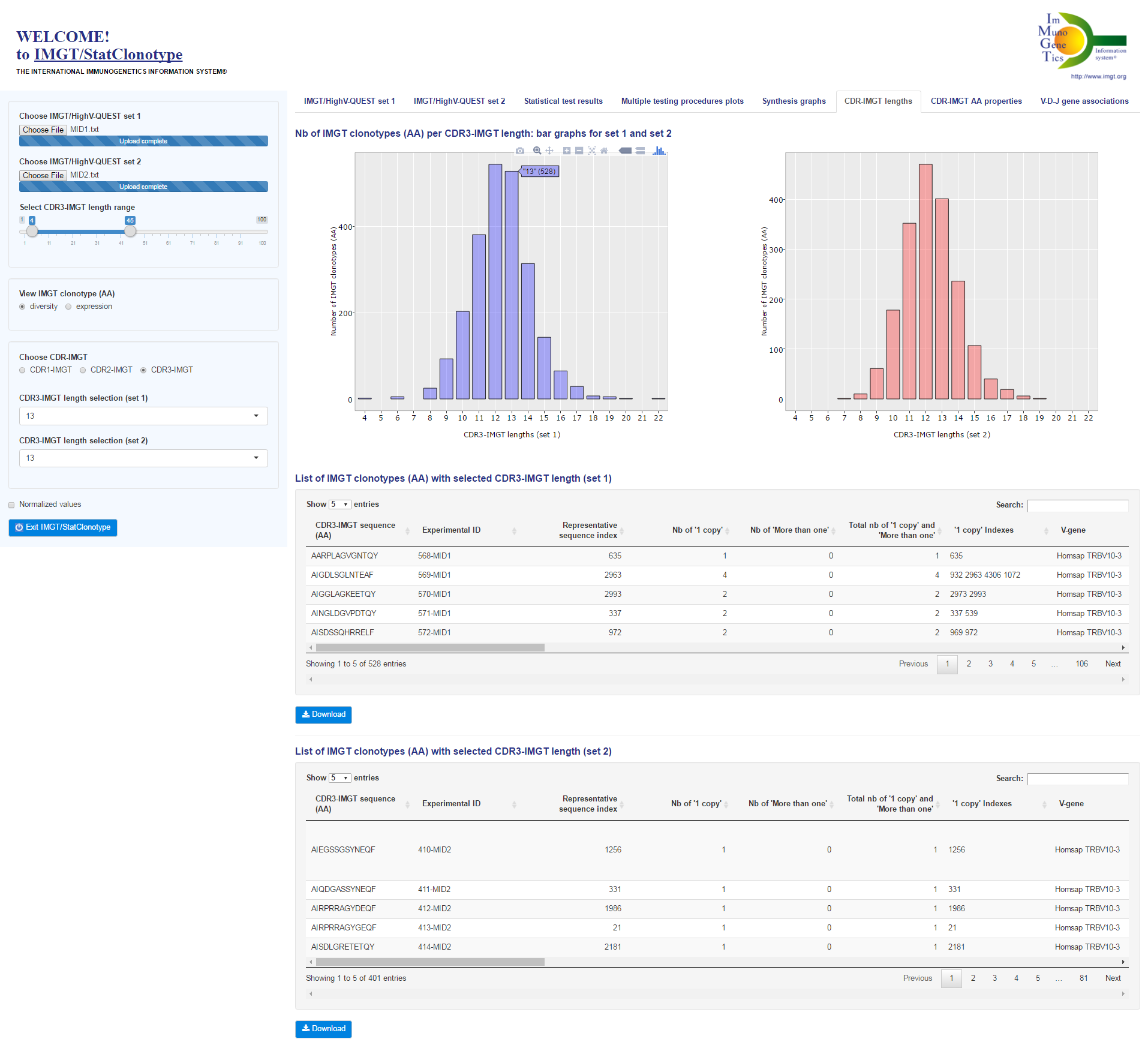
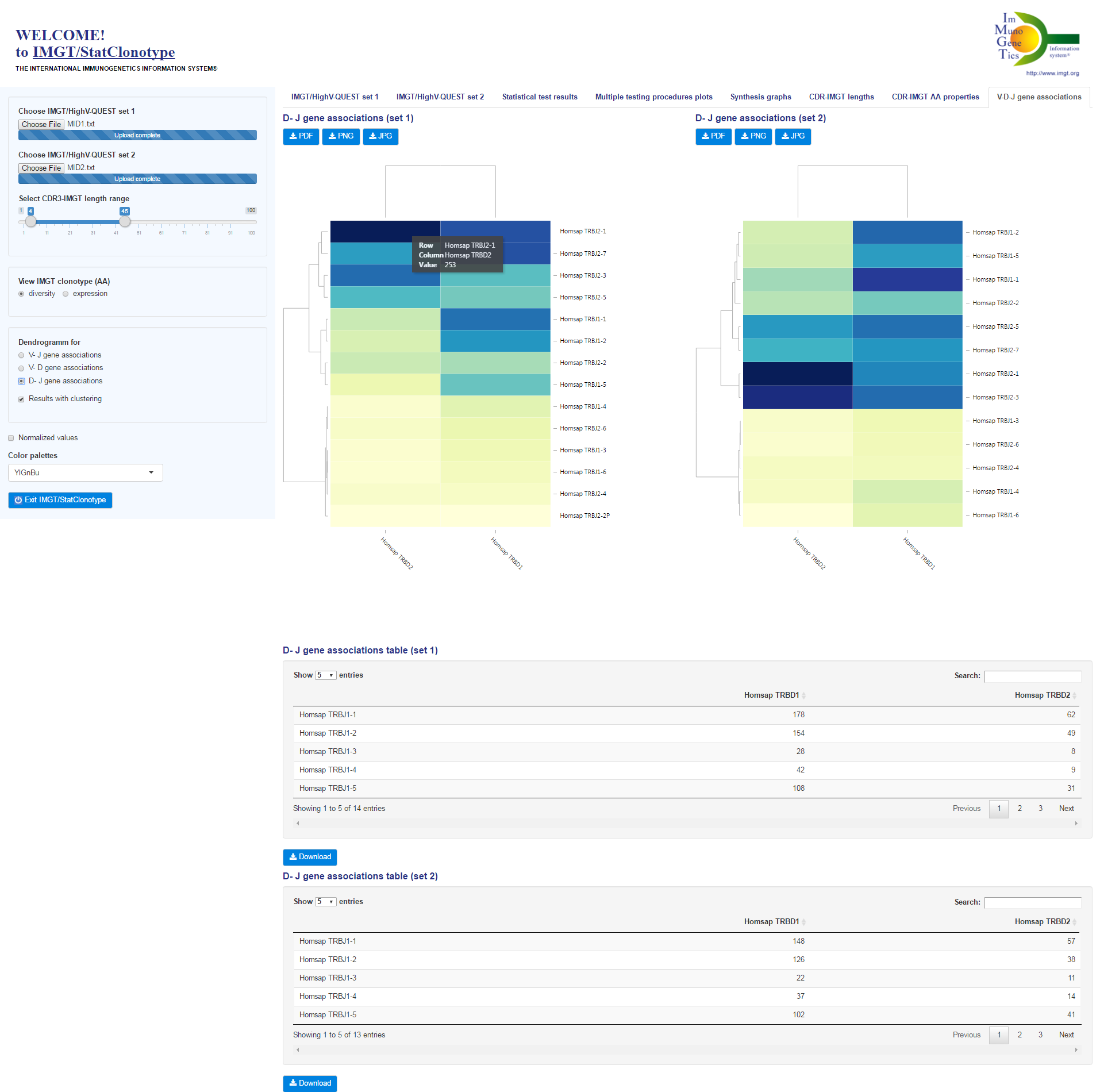
| IMGT/StatClonotype implementation and developments |
The IMGT/StatClonotype tool was developed by Safa Aouinti (IMGT, LIGM, Montpellier, France) using R software (version 3.3.0) and Shiny framework [11].
Interactive graphs are generated using ggplot2 [13] with plotly [14] and d3heatmap [15] packages.
This work was granted access to the HPC resources of CINES under the allocation 2014-036029 made by GENCI (Grand Equipement National de Calcul Intensif) and to Très Grand Centre de Calcul
(TGCC) of the Commissariat à l'Energie Atomique et aux Energies Alternatives
(CEA) under the allocation 036029 (2010-2016) made by GENCI
(Grand Equipement National de Calcul Intensif).
| IMGT/StatClonotype upgrades and versions |
The tab 'Multiple testing procedures plots' now displays interactive scatter plots and interactive line graphs with:
| 'IMGTStatClonotype' R package update |
 remove.packages("IMGTStatClonotype") # To avoid any conflict between two versions previously installed, please retype this line until this message is displayed in the R console "Error in remove.packages: there is no package called 'IMGTStatClonotype'"
remove.packages("IMGTStatClonotype") # To avoid any conflict between two versions previously installed, please retype this line until this message is displayed in the R console "Error in remove.packages: there is no package called 'IMGTStatClonotype'" wspace <- getwd() # Press ENTER
wspace <- getwd() # Press ENTER  des =paste(wspace,"/IMGTStatClonotype_1.0.4.tar.gz",sep="") # Press ENTER
des =paste(wspace,"/IMGTStatClonotype_1.0.4.tar.gz",sep="") # Press ENTER  download.file("https://www.imgt.org/download/StatClonotype/IMGTStatClonotype_1.0.4.tar.gz",destfile=des) # Press ENTER
download.file("https://www.imgt.org/download/StatClonotype/IMGTStatClonotype_1.0.4.tar.gz",destfile=des) # Press ENTER  install.packages("IMGTStatClonotype_1.0.4.tar.gz", repos=NULL) # Press ENTER
install.packages("IMGTStatClonotype_1.0.4.tar.gz", repos=NULL) # Press ENTER  install.packages("colourpicker") # To install the new dependency R package "colourpicker"
install.packages("colourpicker") # To install the new dependency R package "colourpicker"  library(IMGTStatClonotype) # Press ENTER
library(IMGTStatClonotype) # Press ENTER  launch() # Press ENTER
launch() # Press ENTER