
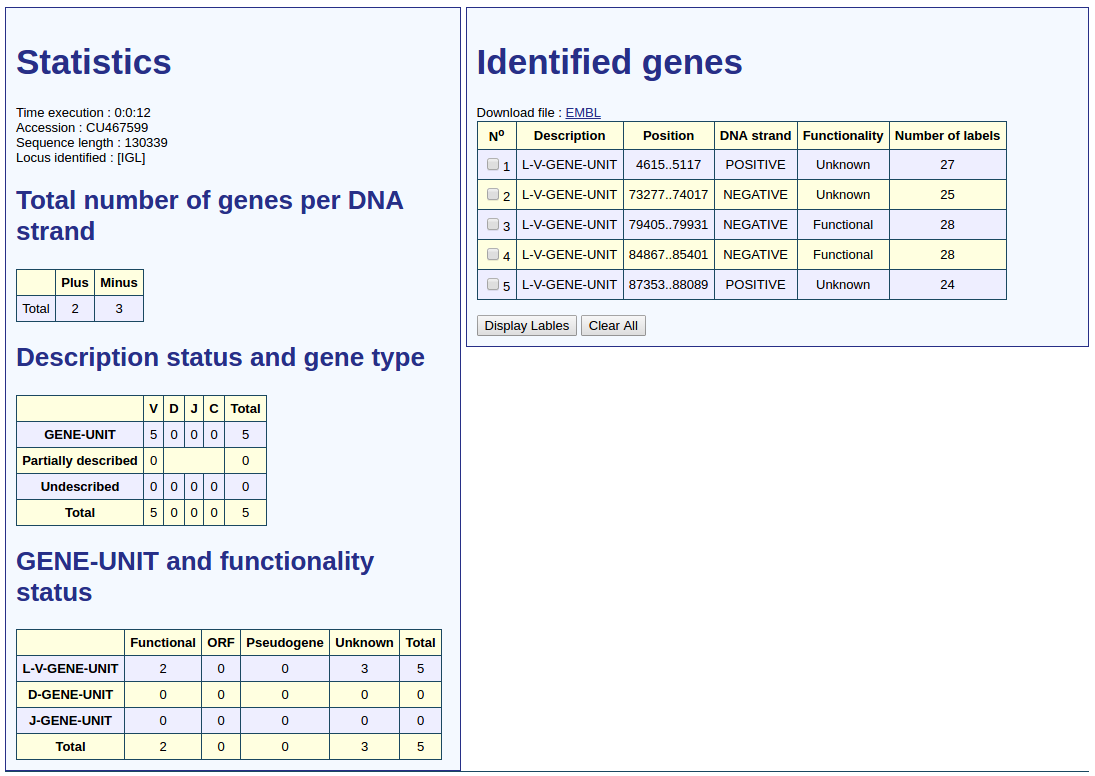
When the automatic annotation of the sequence is completed, the execution time, length and accession number of the analyzed sequence are shown. The results of statistical analysis are divided into three tables. The first provides the total number of genes by DNA strands. The second gives the number of gene depending on the type of gene and description status from the most reliable to the less reliable.
The V gene has the "V undescribed", "V partially described" and "described L-V-GENE-UNIT" status. In the case that less than 2 short motifs are found, the "V undescribed" status is attributed to the gene. This shows that no long motif where delimited from short motifs because 2 short motifs are required to delimit a long motif. In the case that more than 1 short motifs are found but the V-REGION is not described the "V partially described" status is assigned to the gene. This indicates that IMGT/V-QUEST was not used or did not provide results. Long motif may be delimited from short as 2 shot motifs are at least found. In the case that more than 1 short motif are found and the V-REGION is described the "described L-V-GENE-UNIT" status is assigned to the gene.
The D gene has the "D undescribed" and "described D-GENE-UNIT" status. The criteria to assign the "D undescribed" status is the same as for V gene. In the case that more than 1 short motif have been found the "described D-GENE-UNIT" status is assigned to the gene.
The J gene has the "J undescribed" and "described J-GENE-UNIT" status. The criteria to assign the "J undescribed" status is the same as for V and D gene. The criterion to assign the "described J-GENE-UNIT" status is the same as for a D gene.
The C gene has the "C undescribed" and "described C-GENE-UNIT" status. The criteria to assign the "C undescribed" status is the same as for V, D and J gene. The criterion to assign the "described C-GENE-UNIT" status is the same as for D and J gene.
The third allocates the number of genes whose description status is the most reliable, according to their functionality and type.
The genes identified are displayed in order of appearance in the DNA sequence from the 5' to 3' end.
These genes are detailed by 5 columns that provide information on their description status, the gene unit positions in the sequence, the DNA strands, the functionalities and the number of labels describing the gene units.
The results can be exported in EMBL format.
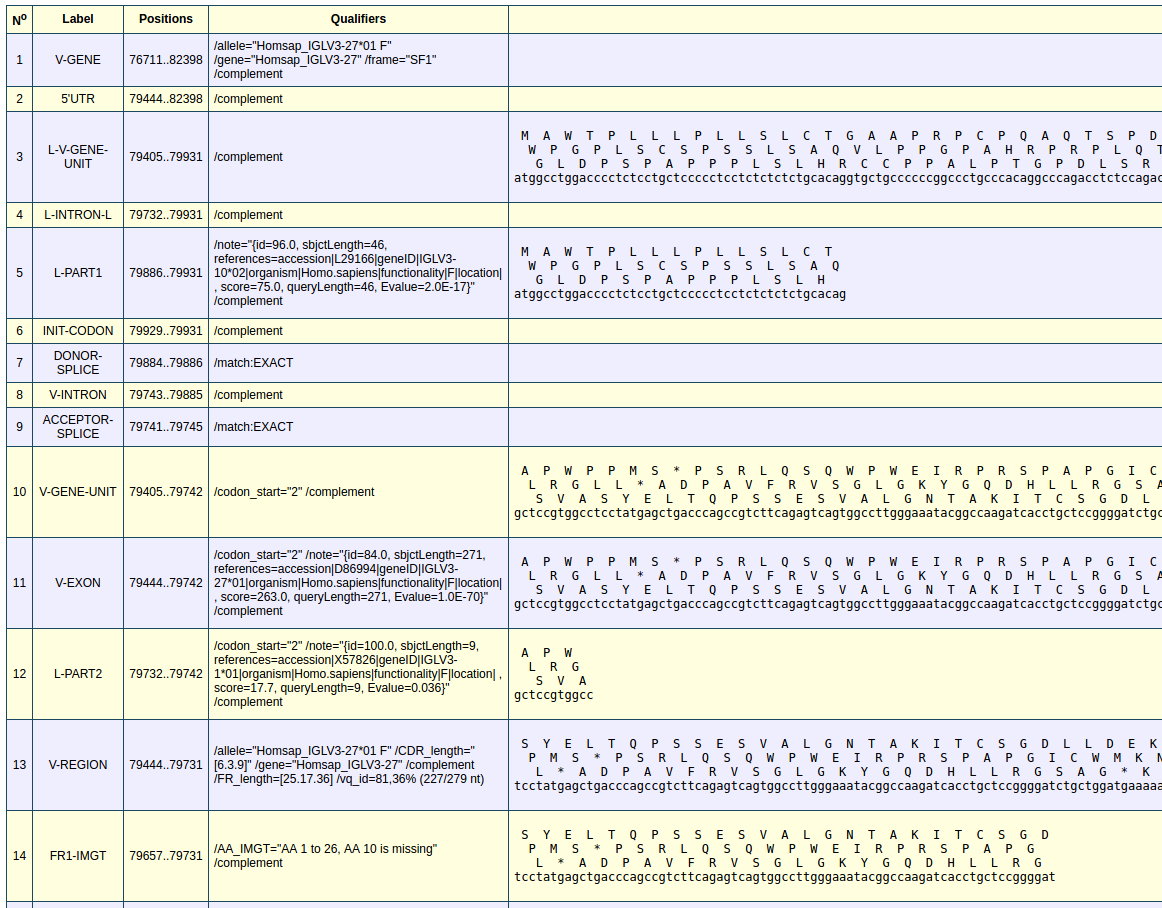
The genes identified can be selected to display all their labels and their characteristics. Here I have selected the V gene. The positions of the labels are provided, the qualifier contain the functionality and other information. The 3 open reading frames of the amino acid sequences are provided with the nucleotide sequence. All the labels including in the V-REGION are provided by IMGT/V-QUEST.
Labels are ordered according to their starting position and nucleotide length. A label that has a starting position upstream of another in the DNA sequence will be displayed above the label. If the two labels shared the same starting positions, the label with the longest nucleotide sequence is displayed above the other label.