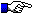 The hinge exon(s) are shown at the end of the protein display.
The hinge exon(s) are shown at the end of the protein display.The IGHC protein display numbering is according to the IMGT unique numbering for C-DOMAIN and C-LIKE-DOMAIN.
Only the *01 allele of each functional, ORF and in-frame pseudogenes C-REGION is shown.
N (Asn, asparagine) of potential N-glycosylation sites (NXS/T, where X is different from P), (N-linked glycosylation) is shown is green (site is underlined in CHS and in pages edited before 14/10/2009).
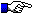 The hinge exon(s) are shown at the end of the protein display.
The hinge exon(s) are shown at the end of the protein display.
_____________________________________________________________________________________________________________________________________________________________________________________________
IGHC
genes
_____________________________________________________________________________________________________________________________________________________________________________________________
A AB B BC C CD D DE E EF F FG G
--------------> ----------> ------> -------> -----------> -------> ---------->
1 10 15 16 20 23 30 36 39 41 45 77 84 85 89 96 97 104 110 118 121 130 140 150
7654321|........|....|123|...|...... ...|.....| |.....|1234567|......|12345677654321|..........|12|....... .....|....... ..|.........|.........|.........|
CH1
X53702 ,IGHA1 (A)SPTSPKVFPLSLDSNPQ....DGNVVVACLVQ GFFPQ.EPLS VTWSESGQGV....TATNFPPSQDASG.....DLYTTSSQLTLPATQC..LAGKSVTC HVKHY...TNPSQ DVTVPCR
X53706 ,IGHA2 (A)SPTSPKVFPLSLDSNPQ....DGNVVVACLVQ GFFPQ.EPLS VTWSESGQNV....TATNFPPSQDASG.....DLYTTSSQLTLPATQC..LAGKSVTC HVTHY...TNPSQ DVTVPCR
M15398 ,IGHE (A)PTRSPSLFPLTRCCKNIPSN.ATSVTLGCLAM GYFP..EPVM VTWDAGSLNG....TTMTLPATTLTPS.....GHYATISLLTVSGAW....AKQMFTC RVAHTPSSTDWVD NKTFS
X65284 ,IGHG1 c (A)STKGPSVFPLAPSSKSTS...GGTQALGCLVK DYFP..EPVT VSWNSGALTS....GVHTFPAVLQSS......GLYSLSSVVTVPSSSL...GTQTYIC NVDHKP..SNTKV DKKV
AF300436,IGHG2 AVLQSS......GLYSLSSVVAVPSSNF...GTQTYTC NVDHKP..SNTKV DKTV
AF300434,IGHG3 AVLQSS......GLYSLSSVVTVPSSSL...GTQTYIC NVDHKP..SNTKV DKRV
AF300432,IGHG4 AVLQSS......GLYSLSSVVTVPSSSL...GTQTYTC NVDHKP..SNTKV DKTV
AF300430,IGHGP AVLQSS......GLYSLSSVVTVPSSSL...GTQTYTC NVDHKP..SNTKV DKRV
CH2
X53702 ,IGHA1 (1) CCHPRLLLHRPALEDLLL..GSEANLTCTLT GLRDA.SGAT FTWSPSSGKS....AVQGPPERDLCG.......CYSVSSVLPGCAEPW..NHGETFTC TAAYPE..SKTPL TANITKS
X53706 ,IGHA2 (1) CCHPRLSLHRPALEDLLL..GSEANLTCTLT GLRDA.SGAT FTWTPPSGKS....AVQGPPERDLCG.......CYSVSSVLPGCAQPW..NHGETFTC TAAHPE..LKNPL TANITKS
M15398 ,IGHE (V)CSRDFTPTVKVLQSSCDGGGHF.PPTIQLLCLVS GYTP..GTIN ITWLEDGQVMD...VDLSTASATQEG......ELASTQSELTLSQKHW..LSDRTYTC QVTYQ....GGTF EDSTKKCA
X61311 ,IGHG1 c (A)PELLGGPSVFLFPPKPKDTLMI.SRTPEVTCVVV DVSHEDPEVK FNWYVDGVEVH...NAKTKPREEQYN......STYRVVSVLTVLHQDW..LNGKEYKC KVSNKA..LPAPI EKTISKAK
AF300436,IGHG2 (A)LPVAGPSVFLFPPKPKDTLMI.SRTPEVTC
AF300434,IGHG3 (A)PELLGGPSVFLFPPKPKDTLMI.TRTPEVTC
AF300432,IGHG4 (A)PEFLGGPSVFLFPPKPKDTLMI.SRTPEVTC
AF300430,IGHGP (A)PELLGGPSVFLFPPKPKDTLMI.SRTPEVTC
CH3
X53702 ,IGHA1 (G)NTFRPEVHLLPPPSEELAL..NELVTLTCLAR GFSP..KDVL VRWLQGSQELPRE.KYLTWASRQEPSQG...TTTFAVTSILRVAAEDW..KKGDTFSC MVGHEA.LPLAFT QKTIDRLA...GKPTHVNVSVVMAEVDGTCY
X53706 ,IGHA2 (G)NTFRPEVHLLPPPSEELAL..NELVTLTCLAR GFSP..KDVL VRWLQGSQELPRE.KYLTWASRQEPSQG...TTTYAVTSILRVAAEDW..KKGDTFSC MVGHEA.LPLAFT QKTIDRLA...GKPTHINVSVVMAEVDGTCY
M15398 ,IGHE (D)SNPRGVSAYLSRPSPFDLFI..RKSPTITCLVV DLAPSKGTVN LTWSRASGKPV...NHSTRKQEKQRN......GTLTVTSTLPVGTRDW..IEGETYQC RVTHPH..LPRAL VRSTTKTS
X61310 ,IGHG1 c (G)QPREPQVYTLPPSRDELT...KNQVSLTCLVK GFYP..SDIA VEWESSGQPEN...NYKTTPPVLDSD......GSFFLYSKLTVDKSRW..QQGNVFSC SVMHEA.LHNHYT QKS
CH4
M15398 ,IGHE (G)PRAAPEVYAFATPEGPGS...RDKRTLACLIQ NFMP..EDIS VQWLHNEVQLPDA.RHSTTQPHKTKG......SGFFVFSRLEVTRAEW..EQKDEFIC RAVHEAASPSQTV QRTVSVNP...GK
Hinge
X53702 ,IGHA1 (V)PSTPCPPTPSTPPTPSPS
X53706 ,IGHA2 (V)PPPPP
X65285 ,IGHG1 c (E)PKSCDTTHTCPPCA
AF300436,IGHG2 (E)RKCCVECPPCP
AF300434,IGHG3 [H1] (E)LKTPLGDTTHTCPRCP [H2] (E)PKSCDTPPPCPRCP
AF300432,IGHG4 (E)SKYGPQCPSCP
AF300430,IGHGP (E)PKTPCCDTTHTCPPCA
IMGT note:
| (1) | In the IGHA gene, the hinge (H) and CH2 are joined in a single exon. |
Created: 12/11/2002
Author: Nathalie Bosc