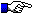 The hinge exon(s) are shown at the end of the protein display.
The hinge exon(s) are shown at the end of the protein display.The IGHC protein display numbering is according to the IMGT unique numbering for C-DOMAIN and C-LIKE-DOMAIN.
Letters in red correspond to amino acids which are polymorphic in the other alleles.
For C-REGIONs, letters in bold correspond to additional positions in the IMGT unique numbering.
For C-REGIONs, letters between parentheses correspond to amino acids resulting from the splicing.
N (Asn, asparagine) of potential N-glycosylation sites (NXS/T, where X is different from P), (N-linked glycosylation) is shown is green (site is underlined in CHS and in pages edited before 14/10/2009).
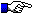 The hinge exon(s) are shown at the end of the protein display.
The hinge exon(s) are shown at the end of the protein display.
_____________________________________________________________________________________________________________________________________________________________________________________________
IGHC
genes
_____________________________________________________________________________________________________________________________________________________________________________________________
A AB B BC C CD D DE E EF F FG G CHS
--------------> ----------> ------> -------> -----------> -------> ---------->
1 10 15 16 20 23 30 36 39 41 45 77 84 85 89 96 97 104 110 118 121 130 140 150
7654321|........|....|123|...|...... ...|.....| |.....|1234567|......|12345677654321|..........|12|....... .....|....... ..|.........|.........|.........|
CH1
X53708 ,IGHA1 (A)SPTSPKVFPLSLCSTQP....DGNVVVACLVQ GFFPQ.EPLS VTWSESGQGV....TATNFPPSQDASG.....GLYTTSSQLTLPATEC..LAGKSVTC HVEHY...TNPSQ DVTVPCR
X53709 ,IGHA2 (A)SPTSPKVFPLSLDSTQP....DGNVVVACLVQ GFFPQ.EPLN VTWSESGQNV....TATNFPPSQDASG.....GLYTTSSQLTLPATEC..LAGKSVTC HVEHY...TNPSQ DVTVPCR
CH2
X53708 ,IGHA1 (1) CCQPRLSLHRPALEDLLL..GSEANLTCTLT GLRDA.SGVT FTWTPSSGKN....AVQGPSERDLCG.......CYSVSSILPGCAEPW..NHGKTFTC TAAQPE..SETQL TATLSKS
X53709 ,IGHA2 (1) CCQPRLSLQRPALEDLLF..GSEANLTCTLT GLRDA.SGAT FTWTPSSGKN....AVQGLPERDLCG.......CYSVSSILPGCAQPW..NNRETFTC TAEHPE..LKTPL TANITKS
CH3
X53708 ,IGHA1 (G)NTFRPEVHLLPPPSEELAL..NELVTLTCLAR GFSP..EDVL VRWLQGSQELPRE.KYLTWASRQEPSQG...TTTFAVTSILRVAAEDW..KKGDTFSC MVGHEA.LPLAFT QKTIDRLA...GKPTHVNVSVVMAEVDGTCY
X53709 ,IGHA2 (G)KNKFRPEVHLLPPPSEELAL..NELVTLTCLAR GFSP..EDVL VRWLQGSQELPRE.KYLTWASRQEPSQG...TTTFAVTSILRVAAEDW..KKGDTFSC MVGHEA.LPLAFT QKTIDRLA...GKPTHVNVSVVMAEVDGTCY
Hinge
X53708 ,IGHA1*01 (V)PLPTPPHP
X53386 ,IGHA1*02 (A)TPPTPPHP
X53709 ,IGHA2 (A)PPPHP
IMGT note:
| (1) | In the IGHA gene, the hinge (H) and CH2 are joined in a single exon. |
Created: 12/11/2002
Author: Nathalie Bosc