
IMGT/DomainSuperimpose Documentation
IMGT®, the international ImMunoGeneTics information system®

IMGT/DomainSuperimpose is part of IMGT®, the international ImMunoGeneTics information system®, the high-quality integrated information system specialized in immunoglobulins (IG), T cell receptors (TR), major histocompatibility complex (MH) of human and other vertebrates species, immunoglobulin superfamily (IgSF), MH superfamily (MhSF) and related proteins of the immune system (RPI), created in 1989 by Marie-Paule Lefranc (Laboratoire d'ImmunoGénétique Moléculaire, LIGM, Université de Montpellier and CNRS) and on the Web since July 1995.
IMGT/DomainSuperimpose superimposes domains of IG, TR, MH and RPI chains with 3D structures in IMGT/3Dstructure-DB, using the method of the least squares fit for the structural comparison. Structural data are extracted from IMGT/3Dstructure-DB.
The tool provides a custom made structural alignment based on the IMGT unique numbering, calculates the root-mean-square deviation (RMSD) for the two structures to be superimposed and displays the two superimposed domains.
Domain sequences are numbered according to the IMGT unique numbering for V-DOMAIN and V-LIKE-DOMAIN [1], to the IMGT unique numbering for C-DOMAIN and C-LIKE-DOMAIN [2], and/or to the IMGT unique numbering for G-DOMAIN and G-LIKE-DOMAIN[3].
Fitting is performed using the McLachlan algorithm [4] as implemented in the program ProFit (Martin, A.C.R., ProFit).
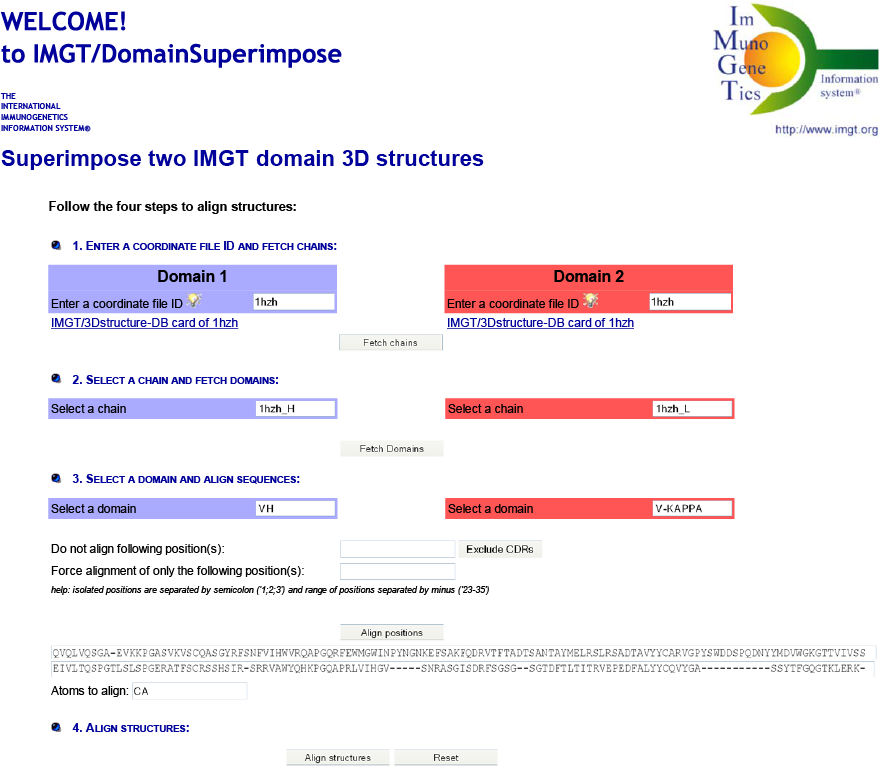
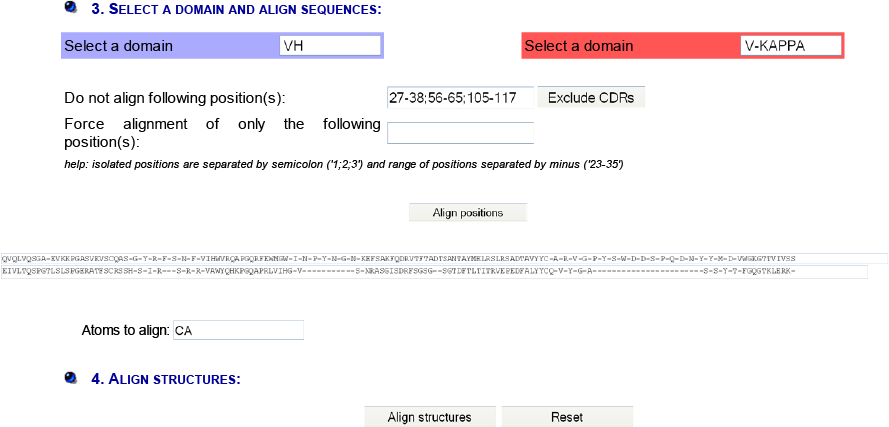
IMGT/DomainSuperimpose allows one:
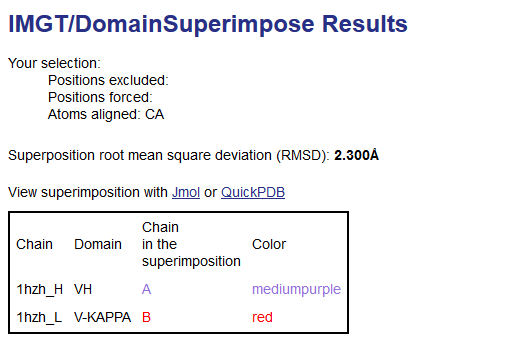
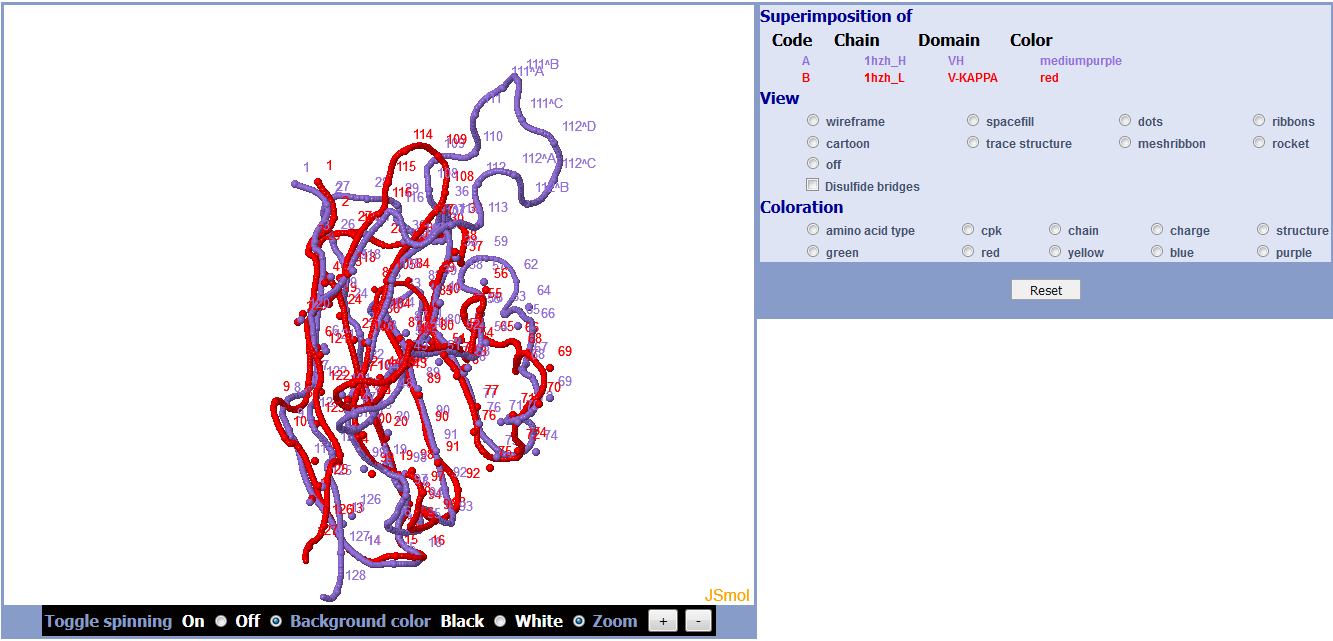
Results show the superposition root mean square deviation (RMSD) in angstrom (Å) and the fitted coordinates for the two domains with chain and color used in the superimposition by JSmol.
| [1] | Lefranc, M.-P., Pommié, C., Ruiz, M., Giudicelli, V., Foulquier, E., Truong, L., Thouvenin-Contet, V. and Lefranc G., IMGT unique numbering for immunoglobulin and T cell receptor variable domains and Ig superfamily V-like domains, Dev Comp Immunol 27:55-77 (2003). PMID:12477501 |
| [2] | Lefranc, M.-P., Pommié, C., Kaas, Q., Duprat, E., Bosc, N., Guiraudou, D., Jean, C., Ruiz, M., Da Piedade, L., Rouard, M., Foulquier, E., Thouvenin, V. and Lefranc G. IMGT unique numbering for immunoglobulin and T cell receptor constant domains and Ig superfamily C-like domains, Dev Comp Immunol 29:185-203 (2005). PMID:15572068 |
| [3] | Lefranc, M.-P., Duprat, E., Kaas, Q., Tranne, M., Thiriot, A. and Lefranc G. (2005) IMGT unique numbering for MHC groove G-DOMAIN and MHC superfamily MhcSF G-LIKE-DOMAIN Dev Comp Immunol 29:917-938. PMID:15936075 |
| [4] | McLachlan, A.D. (1982) Rapid Comparison of Protein Structures Acta Cryst A38: 871-873 |
| [5] | Kaas, Q. and Lefranc, M.-P., IMGT/3Dstructure-DB for immunoglobulin, T cell receptor and MHC structural data, ECCB 2002 |
| [6] | Kaas, Q., Analyse structurale des récepteurs d'antigènes dans IMGT et modélisation moléculaire, Thèse de doctorat, Université Montpellier II, 2005 |
| [7] | Kaas, Q. and Lefranc M.-P. (2005) T cell receptor/peptide/MHC molecular characterization and standardized pMHC contact sites in IMGT/3Dstructure-DB. In Silico Biology 5:0046 |