
IMGT/DomainGapAlign Documentation
IMGT®, the international ImMunoGeneTics information system®

Or>AE000658|TRAV4*01|Homo sapiens|F|V-REGION LAKTTQPISMDSYEGQEVNITCSHNNIATNDYITWYQQFPSQGPRFIIQGYKTKVTNEVA SLFIPADRKSSTLSLPRVSLSDTAVYYCLVGD
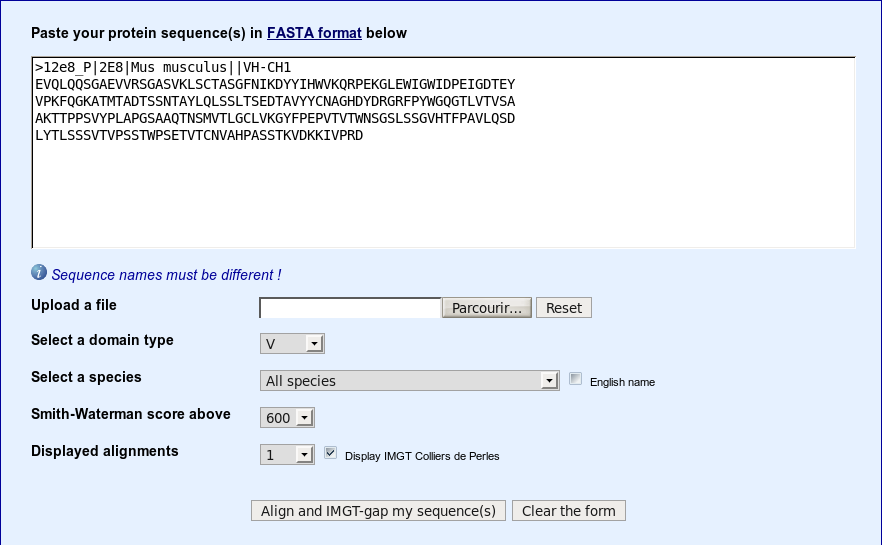
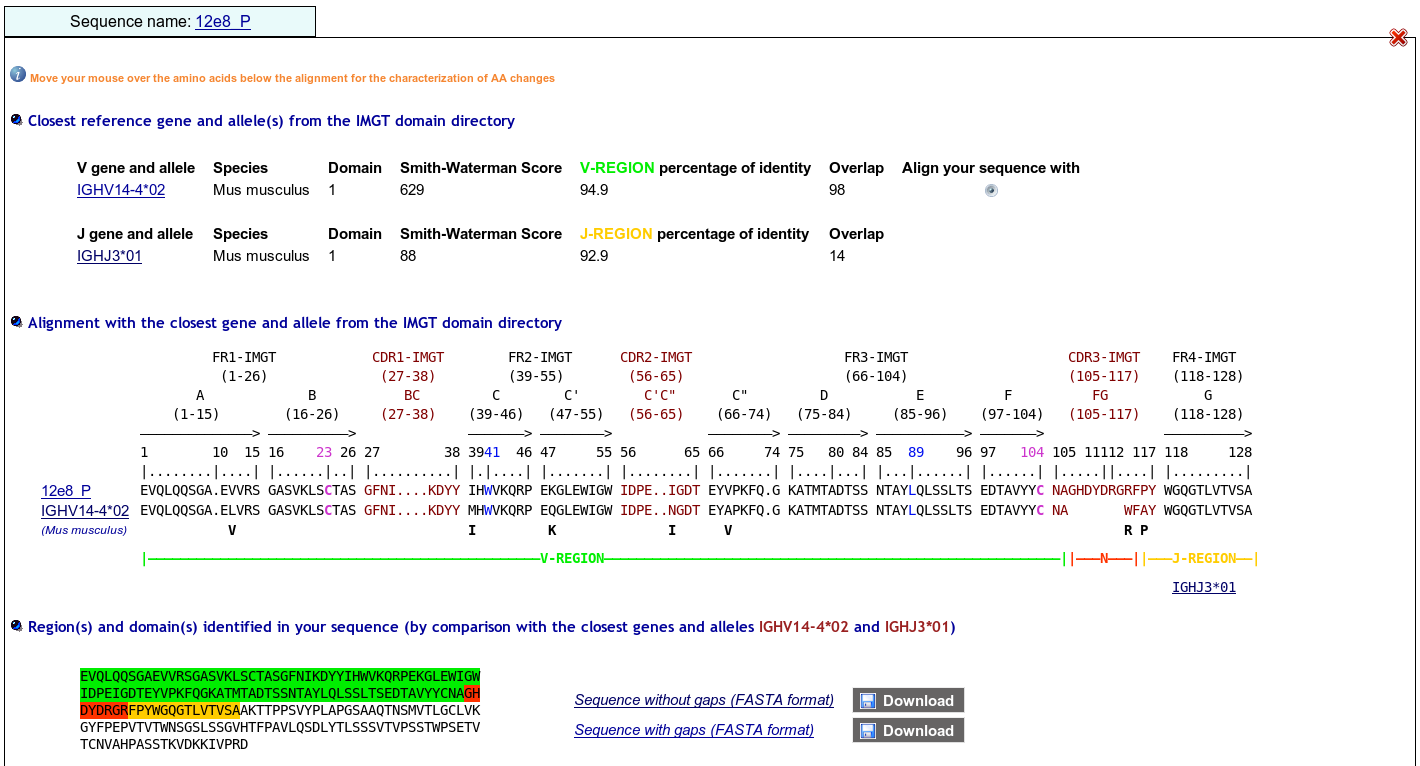
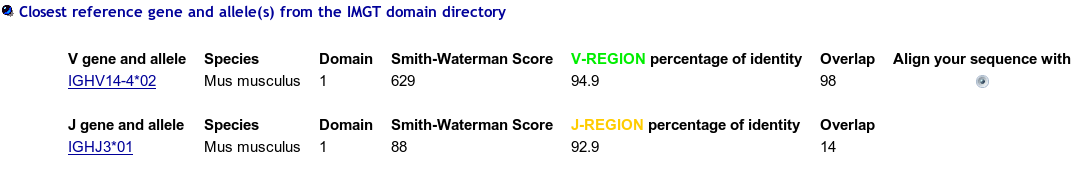
>12e8_P|2E8|Mus musculus||VH-CH1 EVQLQQSGAEVVRSGASVKLSCTASGFNIKDYYIHWVKQRPEKGLEWIGWIDPEIGDTEY VPKFQGKATMTADTSSNTAYLQLSSLTSEDTAVYYCNAGHDYDRGRFPYWGQGTLVTVSA AKTTPPSVYPLAPGSAAQTNSMVTLGCLVKGYFPEPVTVTWNSGSLSSGVHTFPAVLQSD LYTLSSSVTVPSSTWPSETVTCNVAHPASSTKVDKKIVPRD
In IMGT, the 'domain type' concept has three main concepts: the V type (V for variable), the C type (C for constant) and the G type (G for groove).
'V type' identifies the domain type of domains described as V-DOMAIN and V-LIKE-DOMAIN.
'C type' identifies the domain type of domains described as C-DOMAIN and C-LIKE-DOMAIN.
'G type' identifies the domain type of domains described as G-DOMAIN and G-LIKE-DOMAIN.
Select the species in the drop-down list.
Select the minimum or maximum Smith-Waterman score for the alignments to display (For example, choosing a "Smith-Waterman score above 200" will provide and display alignments whose Smith-Waterman score is superior to 200).
Select the number of alignments to display (Displayed alignments) and tick off the checkbox if you want to display IMGT Colliers de Perles.
| V: V-ALPHA, V-BETA, V-DELTA, V-GAMMA, V-KAPPA, V-LAMBDA, V-LIKE. |
| G: G-ALPHA, G-ALPHA1, G-ALPHA1-LIKE, G-ALPHA2, G-ALPHA2-LIKE, G-BETA. |
| C: C-ALPHA, C-BETA-1, C-BETA-2, C-DELTA, C-GAMMA-1, C-GAMMA-2, C-GAMMA-3, C-GAMMA-4, C-GAMMA-5, C-GAMMA-6, C-KAPPA, C-LAMBDA, C-LIKE. |
IMGT nomenclature is based on the CLASSIFICATION axiom and concepts of IMGT-ONTOLOGY [1]. IMGT gene and allele names were approved by the HUGO Nomenclature Committee (HGNC) and by WHO-IUIS Nomenclature Committee [2]. They are used by Entrez Gene (NCBI), GDB, GeneCards, Ensembl (EBI)...
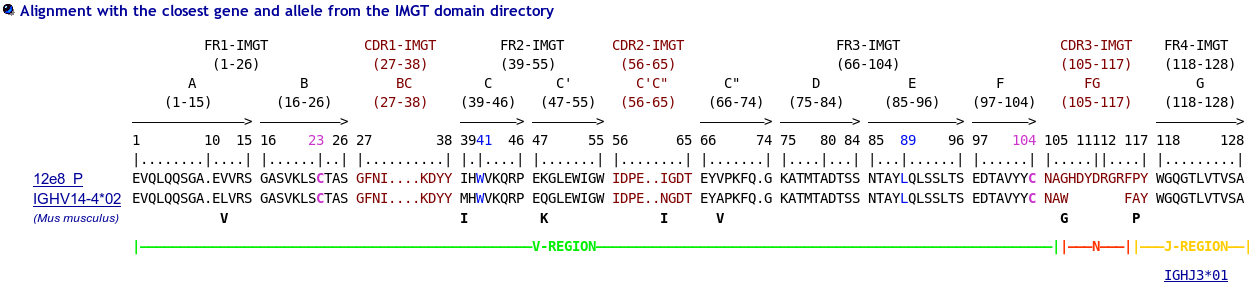

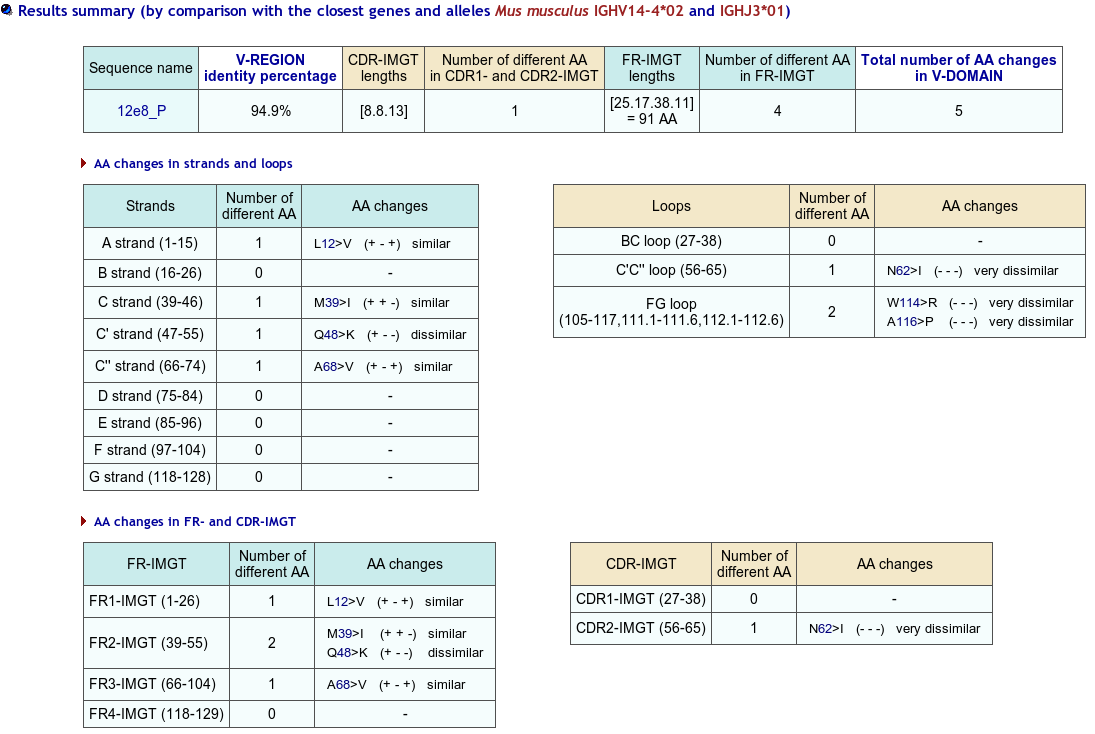
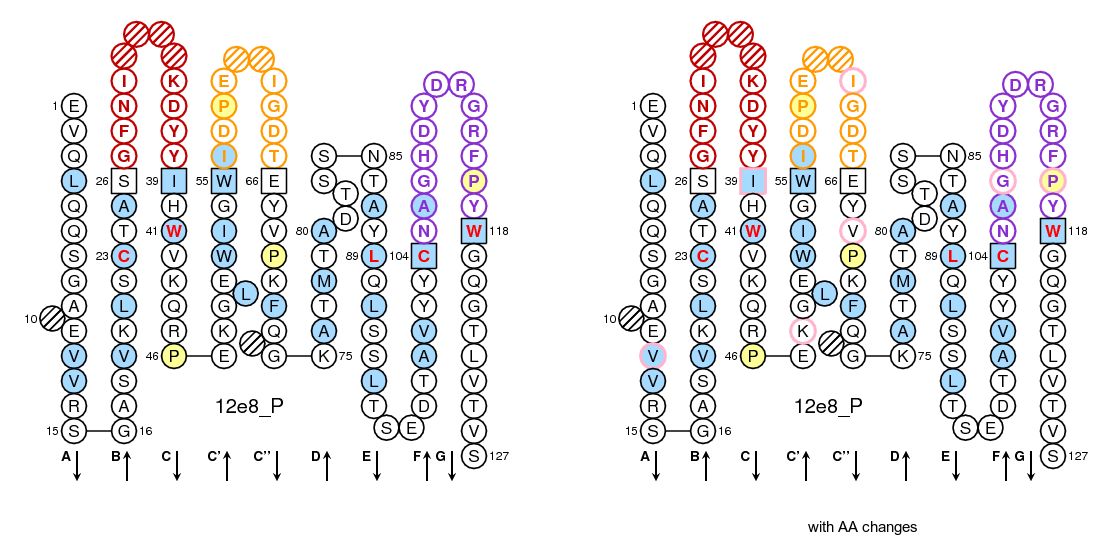
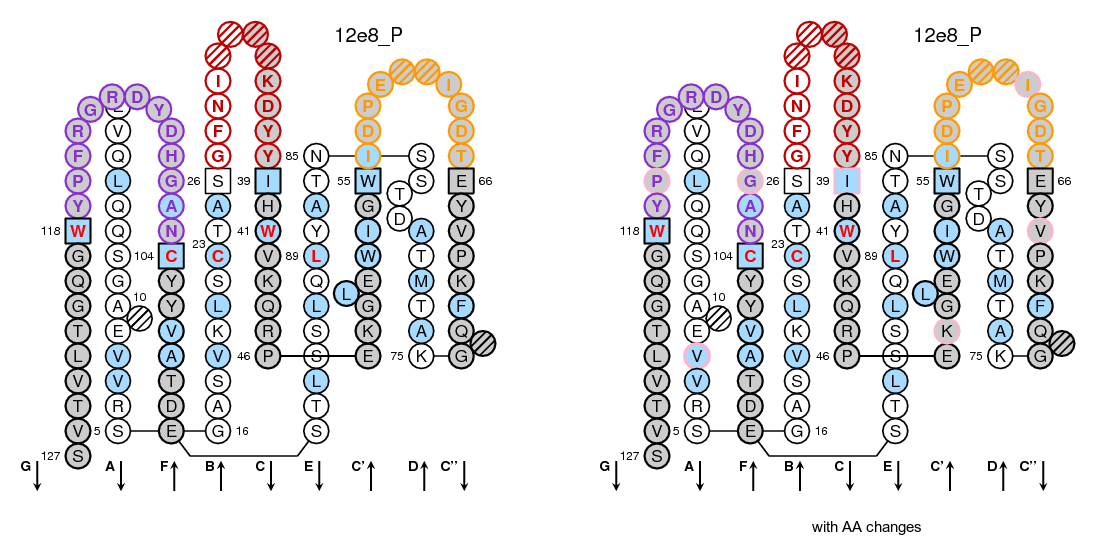
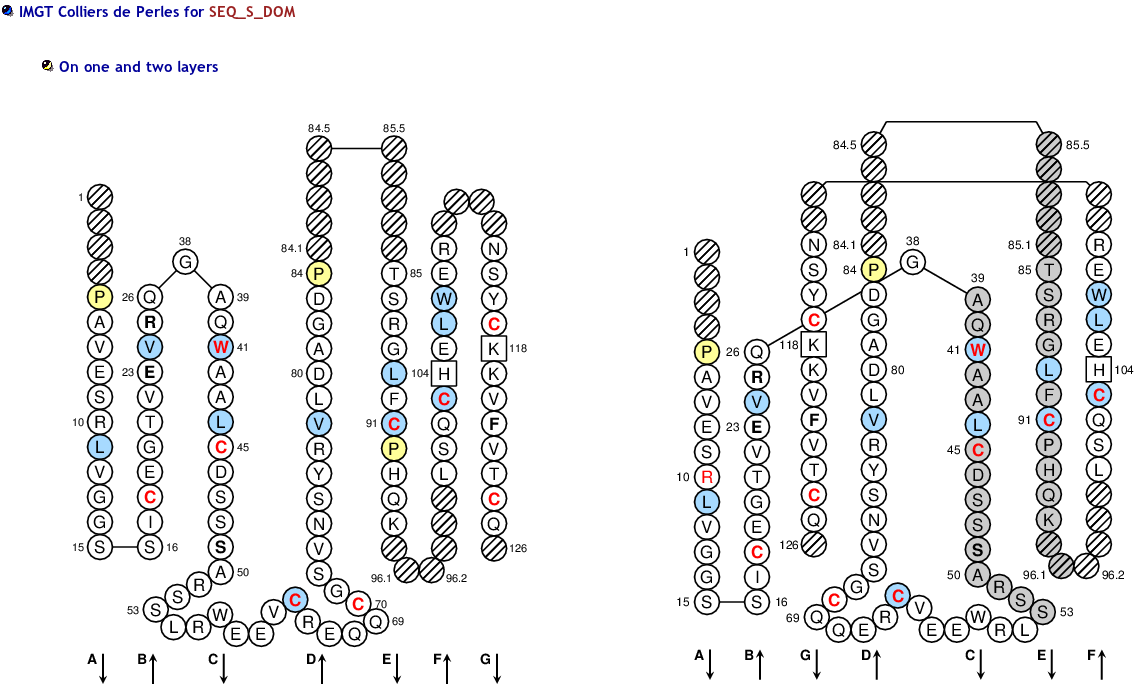
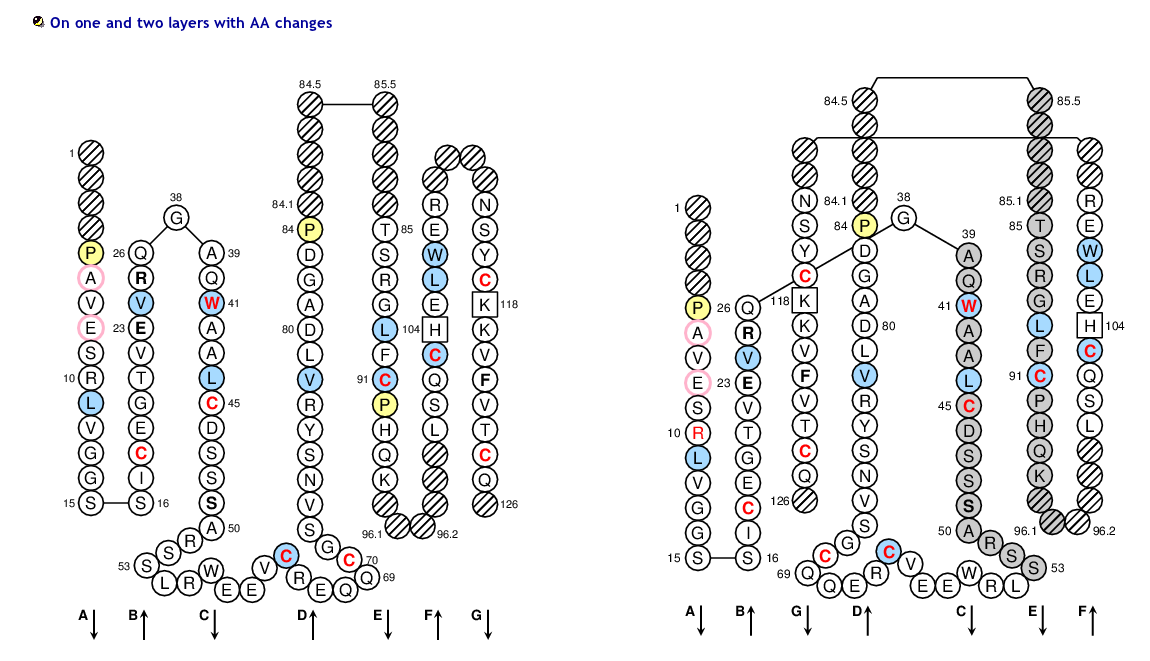
The IMGT/Collier-de-Perles tool allows you to choose the CDR-IMGT color type, the Background color, the Amino acid insertions, the CDR3-IMGT lenght and the domain title:
1) CDR-IMGT color type
- For (IGK,IGL,TRA,TRG), CDR-IMGT regions are colored as follows: CDR1-IMGT (blue), CDR2-IMGT (green) and CDR3-IMGT (greenblue).
- For (RPI,IGH,TRB,TRD), CDR-IMGT regions are colored as follows: CDR1-IMGT (red), CDR2-IMGT (orange) and CDR3-IMGT (purple).
2) Background color
- 50% Hydrophobic positions: These positions shown in blue correspond to hydrophobic amino acids (hydropathy index with positive value) and Tryptophan (W) found at a given position in more than 50% of analysed IG and TR sequences.
- 80% hydropathy, volume and physicochemical classes: These positions correspond to positions defined according to the IMGT classes of the 20 common amino acids for the 'hydropathy', 'volume', 'chemical characteristics' properties
3) Domain sequence
- In the Domain sequence window, seize your gaped sequence with the CDR3 and the J amino acids (at least 9 or 10 amino acids beyond the F or W, of the motif F/WGXG respectively, to get the complete J. See Alignments of Alleles for the IGHJ, IGKJ and IGLJ in IMGT Repertoire.
- add gaps in the CDR3-IMGT. If the CDR3-IMGT is <13 amino acid see IMGT unique numbering for V-DOMAIN and V-LIKE-DOMAIN.
4) In "Your domain title" box (optional), type a clone name (that you would like to be displayed in the figure).
For normal IG and TR domains, you do not need to use the "Amino acid insertions" boxes. These boxes allow to deal with IgSF V-LIKE-DOMAIN and C-LIKE-DOMAIN which have amino acid insertions compared to the IG and TR domains.
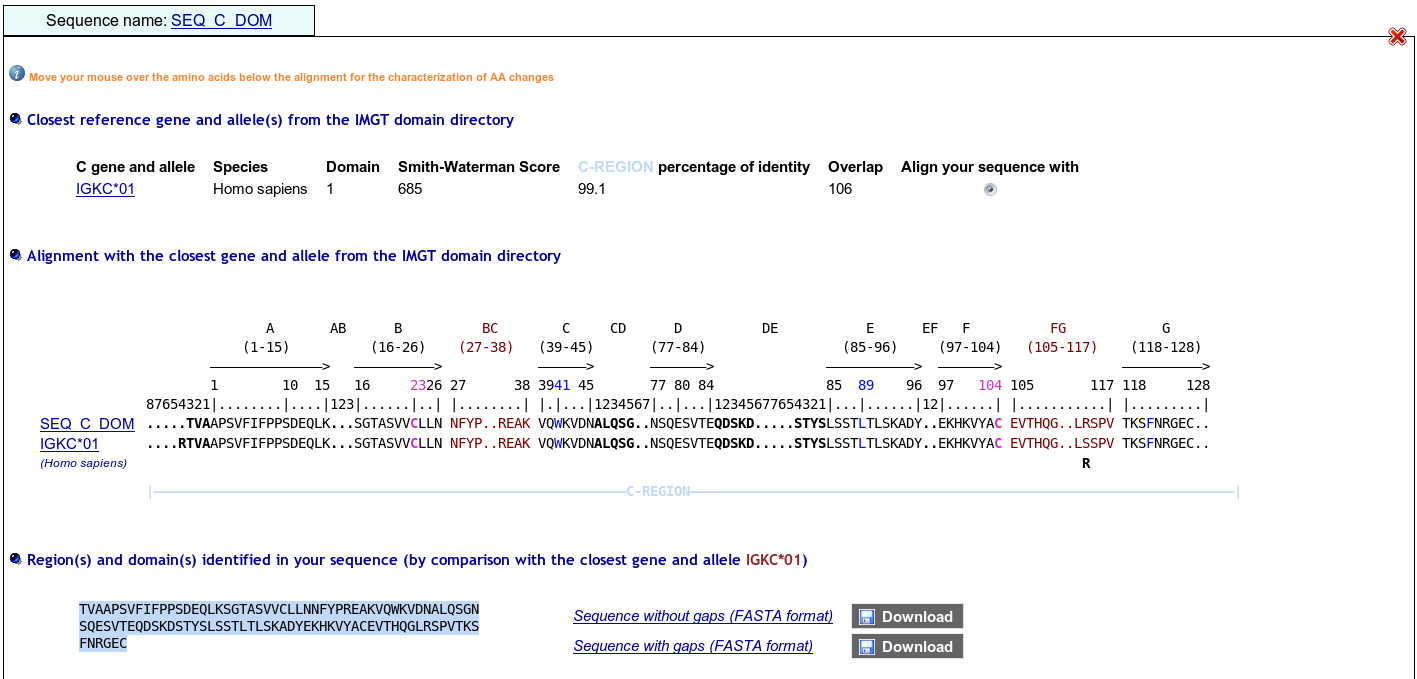
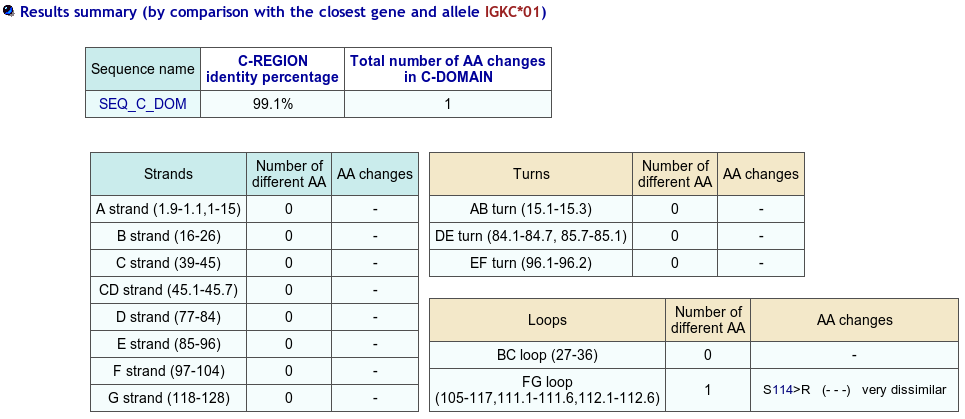
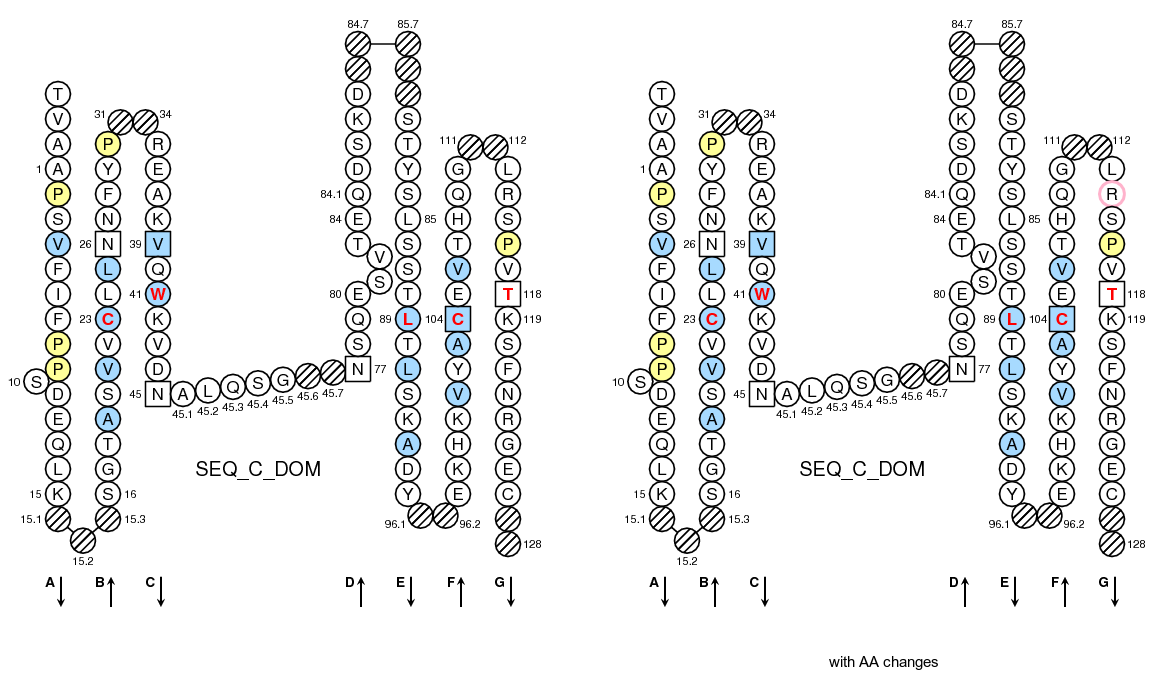
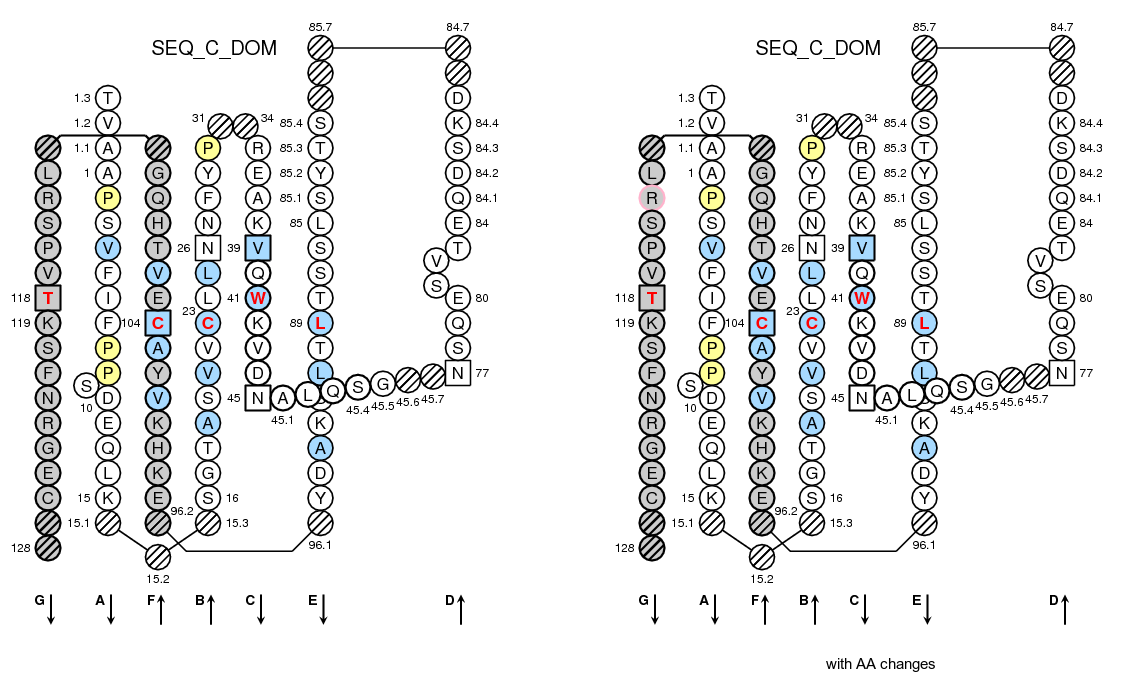
>SEQ_C_DOM TVAAPSVFIFPPSDEQLKSGTASVVCLLNNFYPREAKVQWKVDNALQSGNSQESVTEQDSKDSTYSLSSTLTLSKADYEKHKVYACEVTHQGLRSPVTKSFNRGEC



>pMH75 FRPWFLEYCKSECHFYNTTQRVRLLVRYFYNLELNLRFDSDVGEFRAVTELGQPDAENWNSQPEFLEQKRAEVDTVCRHNYEIFDNFLVPRR
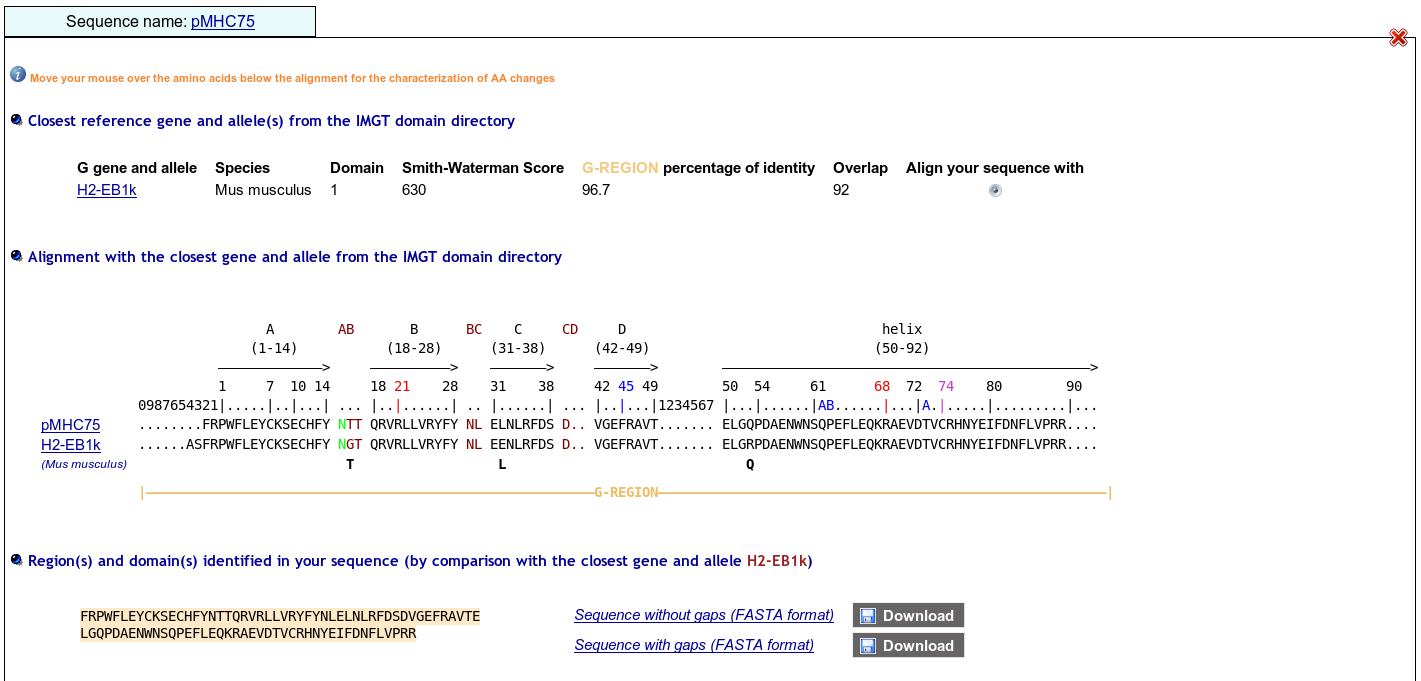

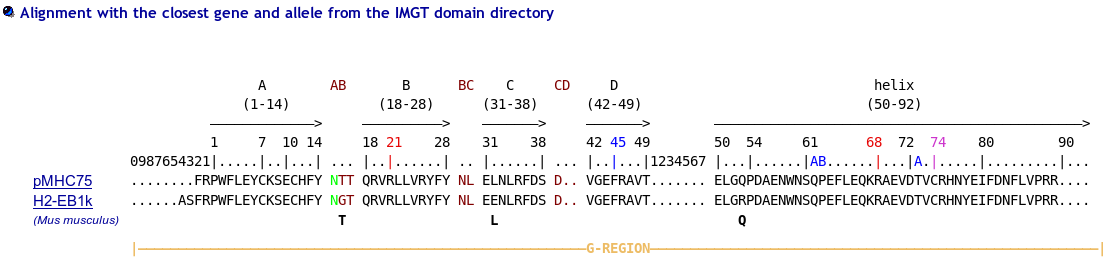

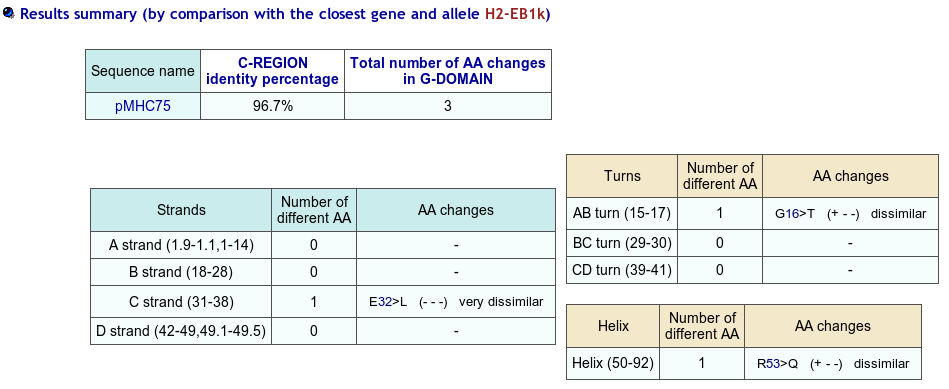
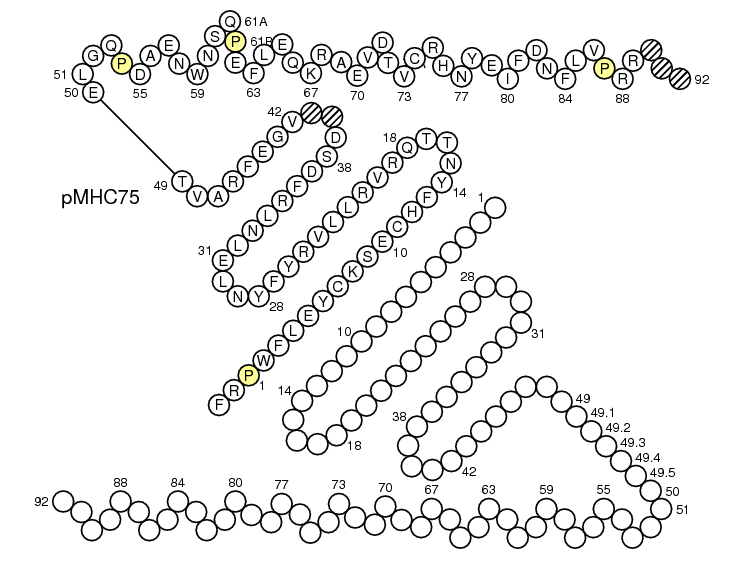
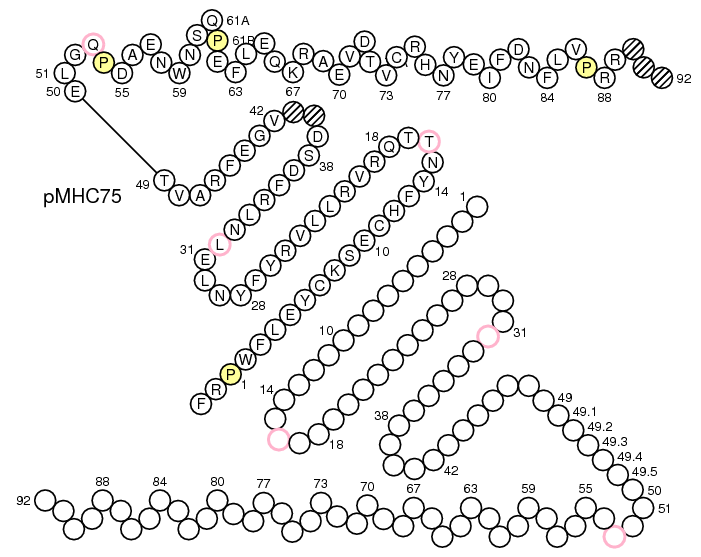
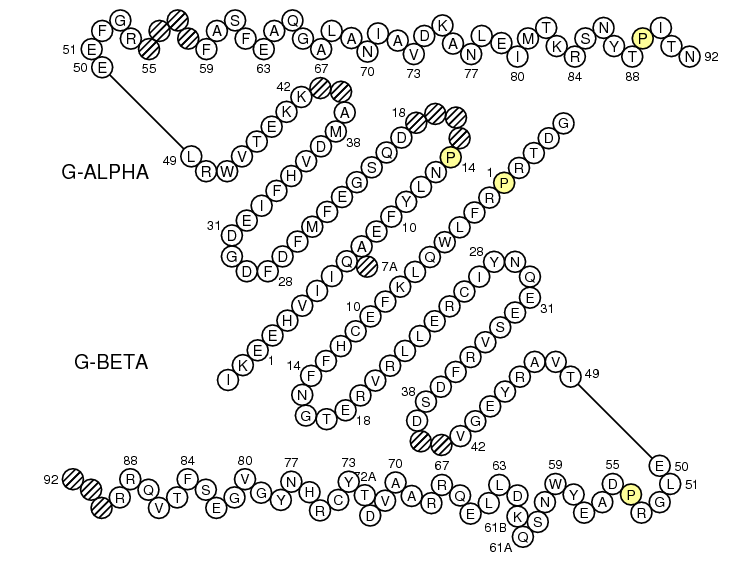
First select Domain type: Groove(G)
1. IMGT/DomainGapAlign steps
Before using IMGT/Colliers-de-Perles for G-domain, align your sequences against the human ones using IMGT/DomainGapAlign to be sure to localize the gaps at the right positions (including the gaps at the beginning of the sequences).
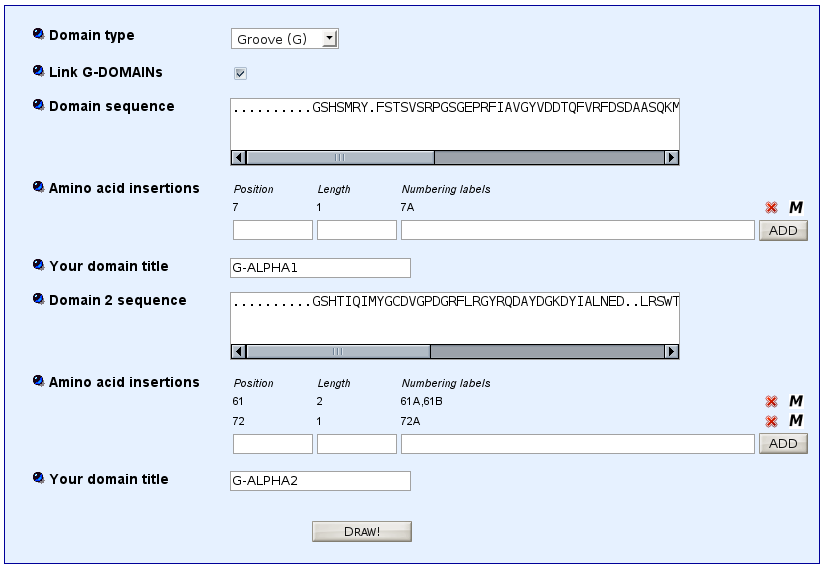
Model for MH class I gaps
>HLA-A*0102 G-ALPHA1 ..........GSHSMRY.FSTSVSRPGSGEPRFIAVGYVDDTQFVRFDSDAASQKMEPRA.......PWIEQEGPEYWDQETRNMKAHSQTDRANLGTLRGYYNQSED..
>HLA-A*0102 G-ALPHA2 ..........GSHTIQIMYGCDVGPDGRFLRGYRQDAYDGKDYIALNED..LRSWTAAD.......MAAQITKRKWEAV.HAAEQRRVYLEGRCVDGLRRYLENGKETLQRT
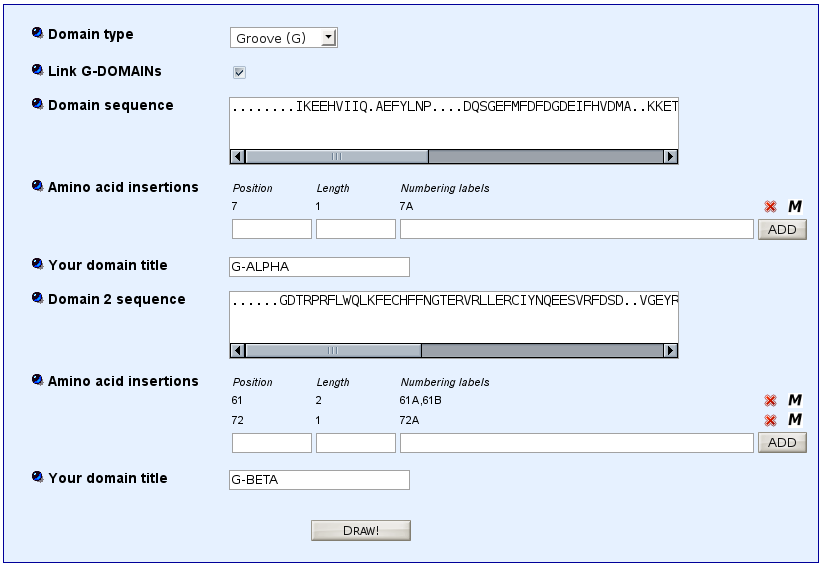
Model for MH class II gaps
>HLA-DRA*0101 G-ALPHA ........IKEEHVIIQ.AEFYLNP....DQSGEFMFDFDGDEIFHVDMA..KKETVWRL.......EEFGR....FASFEAQGALANIAVDKANLEIMTKRSNYTPITN...
>HLA-DRB1*0101 G-BETA ......GDTRPRFLWQLKFECHFFNGTERVRLLERCIYNQEESVRFDSD..VGEYRAVT.......ELGRPDAEYWNSQKDLLEQRRAAVDTYCRHNYGVGESFTVQRR........
Note that IMGT/DomainGapAlign provide more sequences, but be aware that in these alignments the numbering includes gaps for MH1, MH2 and RPI-MH1Like like comparison.
2. IMGT/Collier-de-Perles steps
In Make Your Own IMGT Collier de Perles page (IMGT/Collier-de-Perles), select Domain type: Groove(G).
For MH1, click 'Link G-DOMAINs'.
Paste in the 'Domain 1 sequence' window the sequence of G-ALPHA1 (for MH1) or G-ALPHA (for MH2).
Paste in the 'Domain 2 sequence' window the sequence of G-ALPHA2 (for MH1) or G-BETA (for MH2).
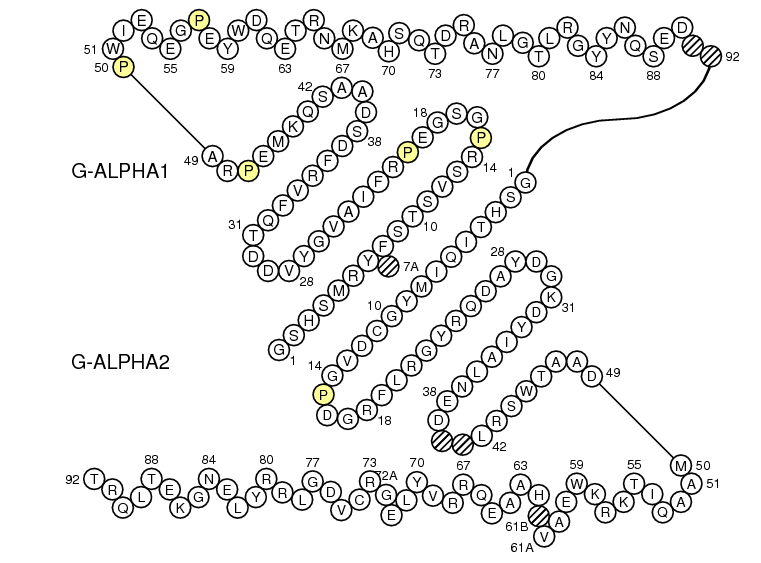
The IMGT "Colliers de Perles" obtained are respectively as follow:
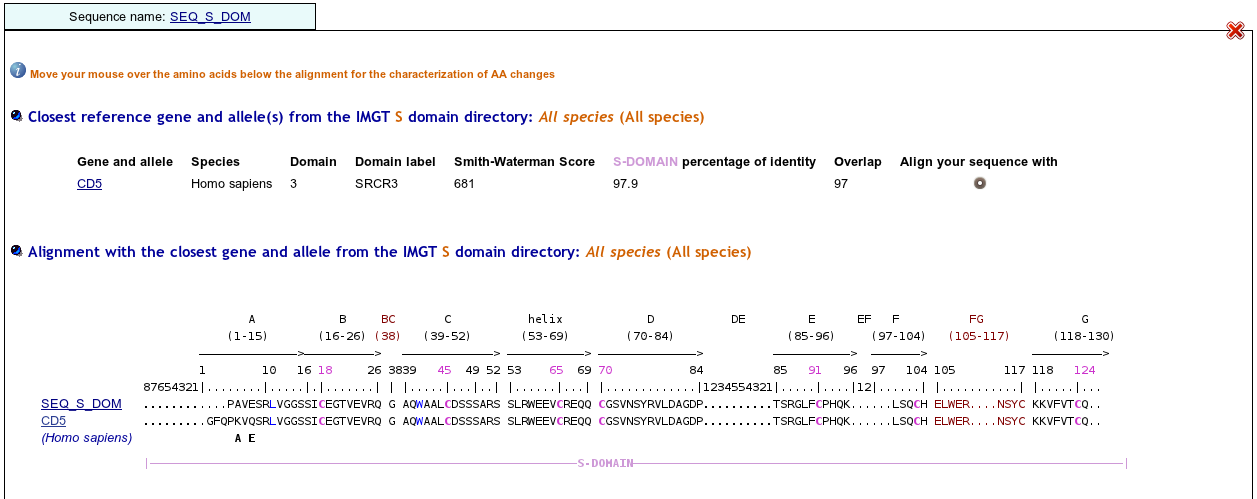
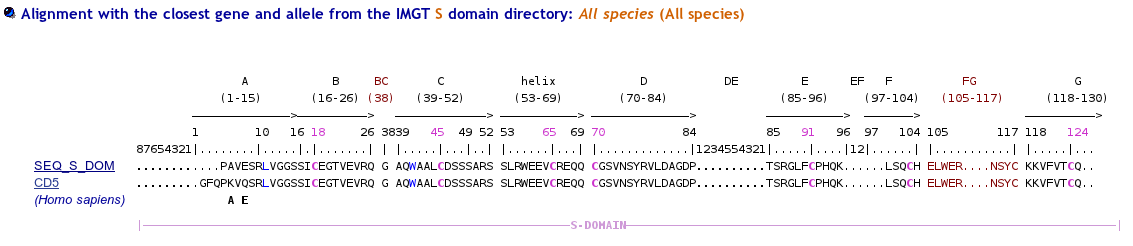
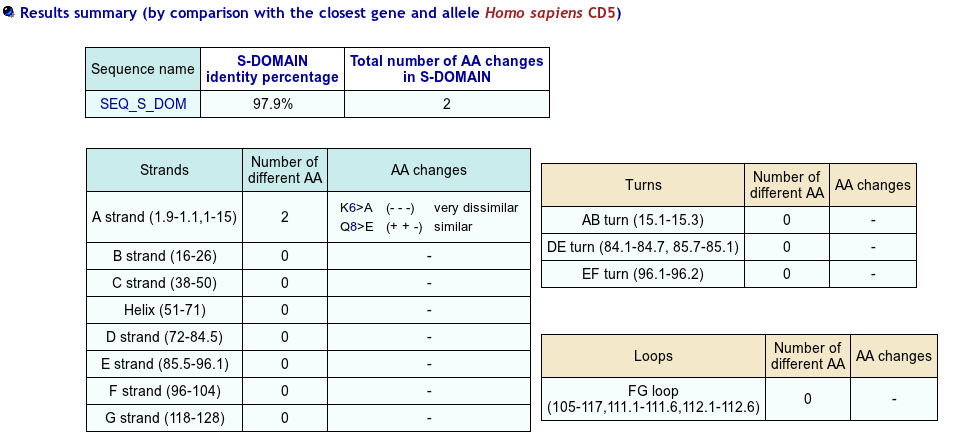
>SEQ_S_DOM PAVESRLVGGSSICEGTVEVRQGAQWAALCDSSSARSSLRWEEVCREQQCGSVNSYRVLDAGDPTSRGLFCPHQKLSQCHELWERNSYCKKVFVTCQD


| [1] | Lefranc, M.-P. , Giudicelli, V., Ginestoux, C., Bosc, N., Folch, G., Guiraudou, D., Jabado-Michaloud, J., Magris, S., Scaviner, D., Thouvenin, V., Combres, K., Girod, D., Jeanjean, S., Protat, C., Yousfi-Monod, M., Duprat, E., Kaas, Q., Pommie, C., Chaume, D. and Lefranc G., IMGT-ONTOLOGY for immunogenetics and immunoinformatics, In Silico Biology 4:17-29 (2004). |
| [2] | Lefranc, M.-P., WHO-IUIS Nomenclature Subcommittee for Immunoglobulins and T cell receptors report, Dev. Comp. Immunol. (in press). |
| [3] | Lefranc, M.-P., Pommié, C., Ruiz, M., Giudicelli, V., Foulquier, E., Truong, L., Thouvenin-Contet, V. and Lefranc G., IMGT unique numbering for immunoglobulin and T cell receptor variable domains and Ig superfamily V-like domains, Dev Comp Immunol 27:55-77 (2003). PMID:12477501. |
| [4] | Lefranc, M.-P., Pommié, C., Kaas, Q., Duprat, E., Bosc, N., Guiraudou, D., Jean, C., Ruiz, M., Da Piedade, L., Rouard, M., Foulquier, E., Thouvenin, V. and Lefranc G., IMGT unique numbering for immunoglobulin and T cell receptor constant domains and Ig superfamily C-like domains, Dev Comp Immunol 29:185-203 (2005). PMID:15572068. |