
IMGT/DomainDisplay Documentation
IMGT®, the international ImMunoGeneTics information system®

IMGT/DomainDisplay is part of IMGT®, the international ImMunoGeneTics information system® http://www.imgt.org [1].
IMGT/DomainDisplay created by LIGM (Université Montpellier, CNRS) is an IMGT tool which provides the display of amino acid sequences from the IMGT domain directory, on the Web since 28 September 2006.
IMGT/DomainDisplay allows queries for IMGT Alignments of domains and polymorphisms of the IgSF and MhcSF superfamilies. Alignments are provided based on the IMGT unique numbering for V-DOMAIN and V-LIKE-DOMAIN [2], C-DOMAIN and C-LIKE-DOMAIN [3], G-DOMAIN and G-LIKE-DOMAIN [4].
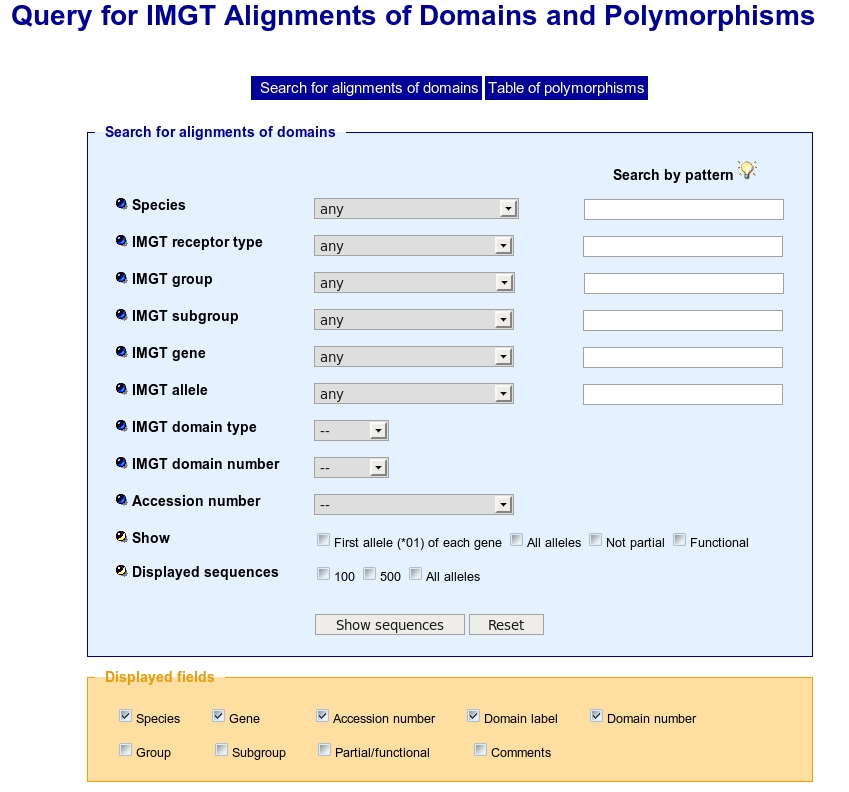
The IMGT/DomainDisplay Query page allows two types of search and offers visualization choice for the results.
This kind of search uses linked select elements. Linked select elements are two or more selects, where choosing a value in one changes the available values in some of the rest. For example, selecting a Species will updates the 'IMGT group' select to only contain group(s) from the Species selected. In the same way, selecting an IMGT group will updates the 'IMGT subgroup' select to only contain subgroup(s) from the IMGT group selected.
Choose in a list a species, and then an: IMGT receptor type, IMGT group, IMGT subgroup, IMGT gene, IMGT allele.
Group, subgroup, gene and allele names are based on the IMGT-ONTOLOGY CLASSIFICATION concept.
Gene:
Allele:
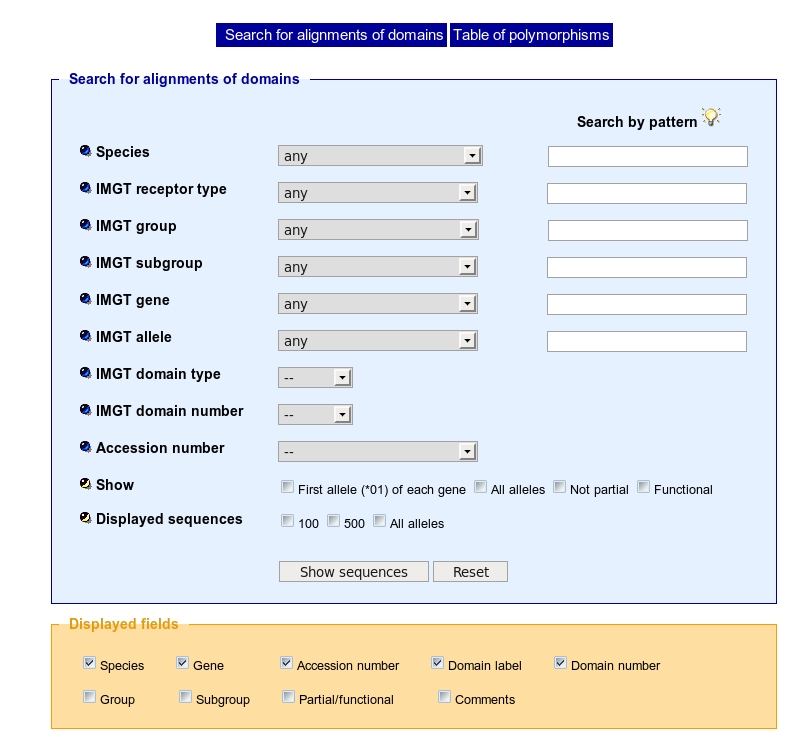
Precise the IMGT domain type, the IMGT domain number, the IMGT functionality or the Accession number to refine research.
You can search IMGT Alignments of Domains by typing a search pattern (for the species, group, subgroup, gene, allele, receptor type and the domain number). The % symbol is used as a wildcard and the comma separate a multiple pattern.
For example, if you type Ho% in the species textarea:
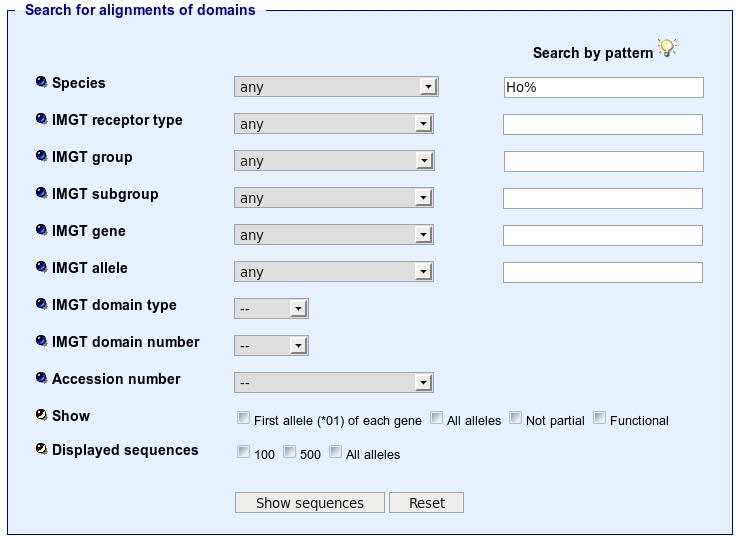
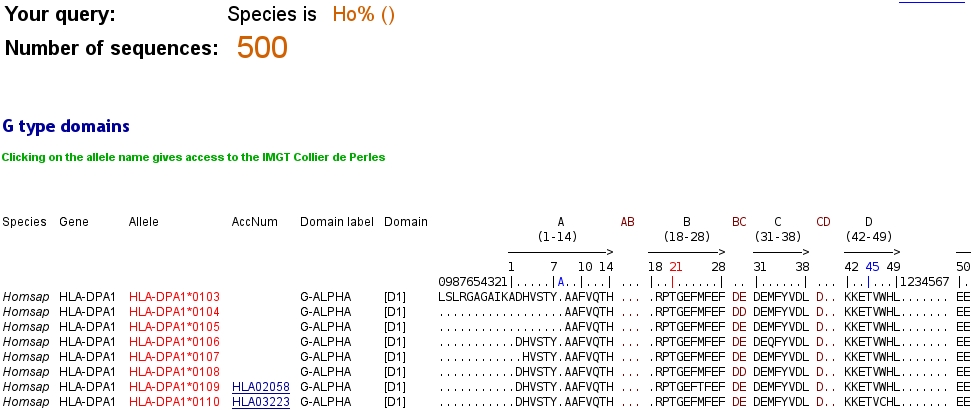
the result page will give you all the IMGT Alignments of Domains for which the species begins by Ho.
At the bottom of the IMGT/DomainDisplay Query page, the user can choose to view the fields that he checked, to seize the maximal number of displayed sequences and to display options.
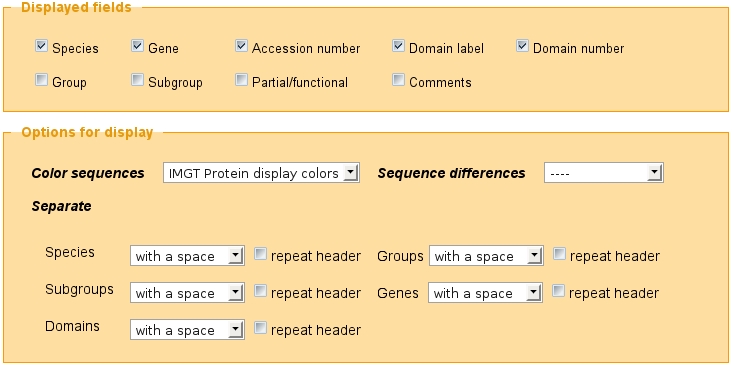 'Displayed fields' menu:
'Displayed fields' menu:
'Displayed options' menu:
The result presentation depends on the visualization choice made at the IMGT/DomainDisplay Query page.
On top of each result page, the visualization choice and the query are recalled, and the number of results is shown.
The domain type is displayed just before the IMGT Alignments of Domains.
The Default overview results page displays the IMGT Alignments of Domains (based on the IMGT color menu for sequence alignments and mutations that satisfy the query, with for each result the following information (See below examples with V type domains, C type domains and J Regions):
V type domains: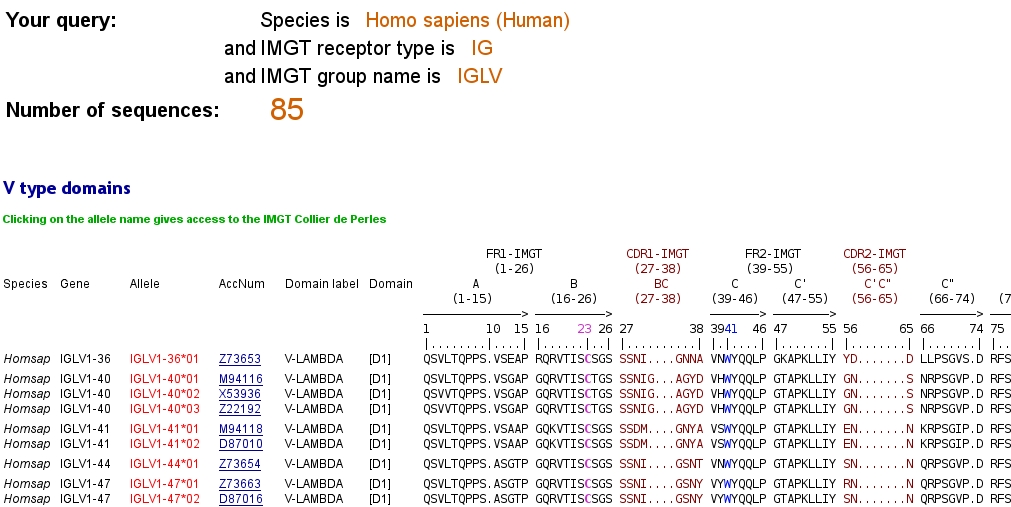
The display of the V type domains is based on the IMGT unique numbering for C-DOMAIN and C-LIKE-DOMAIN.
The 'FR-IMGT or CDR-IMGT sequences' visualization is only for the V type domains.
C type domains: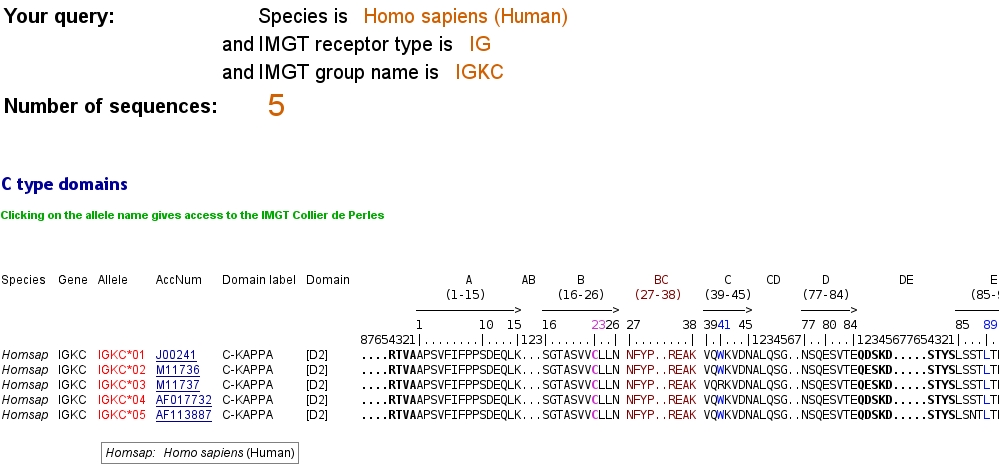
The display of the C type domains is based on the IMGT unique numbering for C-DOMAIN and C-LIKE-DOMAIN.
J REGIONs: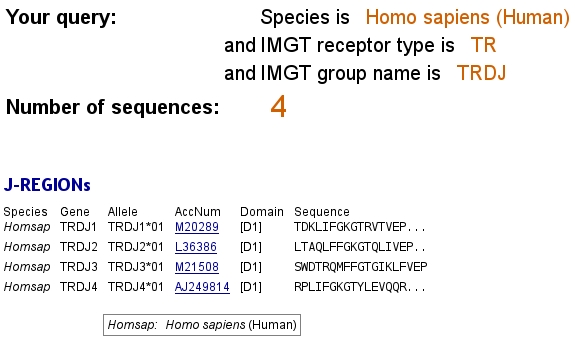
You can display a list of IMGT Alignments of Domains that satisfy the query, with for each entry or chain, the fields of the 'Displayed fields' and 'Display options' menu selected in the visualization choice.
When several alleles are shown, the amino acid changes are indicated in red letters. These polymorphic mutations are reported in Tables of alleles. Currently, the amino acid changes are indicated for V and C type domains. See below the highlighting of polymorphisms for Homo sapiens, IGHV Group.

The first gene identification of the human IG proteins were carried out during a stay in the Laboratoire d'ImmunoGénétique Moléculaire, IGH, CNRS, Montpellier, France, by Dr. Gunilla Norhagen and Dr. Per-Erik Engstrom (May 1998) from the Huddinge University Hospital, Karolinska Institute, Huddinge, Sweden, Dr. Oksana Kravchuk (April-May 1999) from the Research Centre for Medical Genetics, Russian Academy of Medical Science, Moscow, Russia, and Dr. Olga Posukh (April-July 1999, July-October 2000) from the Institute of Cytology and Genetics, Siberian Branch of the Russian Academy of Science, Novosibirsk, Russia.