anti-idiotype anti-idiotype (A48: levan-specific BALB/c myeloma protein ABPC48) anti-idiotype (Ab3) (anti-Neisseria meningitidis polysaccharide C) anti-idiotype (anti-sigma receptor) anti-idiotype (anti-tumor effect) anti-idiotype > cocaine anti-idiotype > cyclosporine (Cs) anti-idiotype HLA class II anti-idiotype [human] anti-idiotype, anti-cancer [human] anti-idiotype, anti-epidermal growth factor receptor
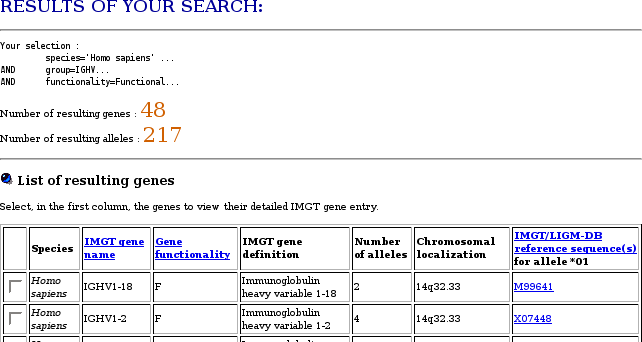
Total number of IG and TR genes Number of functional IG and TR genes Number of genes in the IMGT genome analysis tools
Potential germline repertoires Questions and answers (IMGT Education): Nomenclature and overview of the human immunoglobulin genes Questions and answers (IMGT Education): Nomenclature and overview of the human T cell receptor genes
5' T - C - A - G 3' A - G - T - C
| 5' T - C - A - G 3' A - G - T - C |
} |
5' T - C 3' A- G - T - C - G - A
5' T - C - A - G - C - T 3' A - G - T - C - G - A
>M99641|IGHV1-18*01|Homo sapiens|F|L-PART1+L-PART2|47..92+177..187 atggactggacctggagcatccttttcttggtggcagcaccaacaggtgcccactcc >X60503|IGHV1-18*02|Homo sapiens|F|L-PART1+L-PART2|1..46+131..141 atggactggacctggagcatccttttcttggtggcagcagcaacaggtgcccactcc >X07448|IGHV1-2*01|Homo sapiens|F|L-PART1+L-PART2|126..171+258..268 atggactggacctggaggatcctcttcttggtggcagcagccacaggagcccactcc >X62106|IGHV1-2*02|Homo sapiens|F|L-PART1+L-PART2|21..66+152..162 atggactggacctggaggatcctcttcttggtggcagcagccacaggagcccactcc >X92208|IGHV1-2*03|Homo sapiens|F|L-PART1+L-PART2|18..63+149..159 atggactggacctggaggatcctcttcttggtggcagcagccacaggagcccactcc ...
>M99641|IGHV1-18*01|Homo sapiens|F|L-PART1+L-PART2|47..92+177..187 MDWTWSILFLVAAPTGAHS >X60503|IGHV1-18*02|Homo sapiens|F|L-PART1+L-PART2|1..46+131..141 MDWTWSILFLVAAATGAHS >X07448|IGHV1-2*01|Homo sapiens|F|L-PART1+L-PART2|126..171+258..268 MDWTWRILFLVAAATGAHS >X62106|IGHV1-2*02|Homo sapiens|F|L-PART1+L-PART2|21..66+152..162 MDWTWRILFLVAAATGAHS >X92208|IGHV1-2*03|Homo sapiens|F|L-PART1+L-PART2|18..63+149..159 MDWTWRILFLVAAATGAHS ...
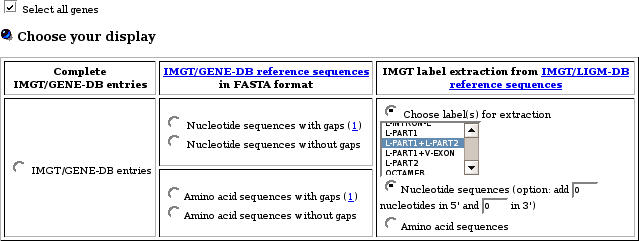
You can access the same tables starting from the IMGT/GENE-DB Query
page:
Click on 'IMGT/GENE-DB direct links for a given gene' (at the bottom of the Query page), and then in the resulting page select the appropriate format:
J-REGION [740..777] gtggacgttcggtggaggcaccaagctggaaatcaaac W T F G G G T K L E I K J-REGION [1094..1132] tgtacacgttcggaggggggaccaagctggaaataaaac Y T F G G G T K L E I K
IMGT label definition: J-REGION: coding region of J-GENE (plus 1 or 2 nucleotide(s) after J-HEPTAMER, if present) or corresponding region in cDNAIn genomic sequences J-HEPTAMER and J-REGION are therefore be contiguous (numbers in green below).
FT J-HEPTAMER 733..739 FT J-REGION 740..777 FT /note="functional" FT /allele="IGKJ1*01" FT /gene="IGKJ1" FT /codon_start=2 FT /translation="WTFGGGTKLEIK" FT J-HEPTAMER 1087..1093 FT J-REGION 1094..1132 FT /note="functional" FT /allele="IGKJ2*01" FT /gene="IGKJ2" FT /codon_start=3 FT /translation="YTFGGGTKLEIK"
Information to be provided
The information to be provided comprises, for each gene:
1) the public accession number of the clone from which the sequence was extracted (or of the NCBI assembly positions and version), with the proposed gene name (provisional), positions start and end in that accession number,
2) the corresponding sequence in FASTA format:
- for V genes: from the beginning of the L-PART1 (atg) to the 3'end of the V-RS
- for D genes: from the 5' end of the 5'D-RS to the 3' end of the 3'D-RS
- for J genes: from the 5' end of the J-RS to the 3'end of the J-REGION
- for C genes: from the 5' end of the first exon to the 3' end (stop codon) of the last exon.
3) the proposed functionality (F, ORF, P), and comments (any comment which can be useful, for example, why a gene is considered 'ORF' or 'P')
4) additionally, for V genes,
- the CDR-IMGT lengths (the germline CDR3-IMGT includes, if present, the one or two nucleotides upstream of the V-HEPTAMER)
- the closest human V-REGION gene and allele with score, percentage of identity and alignment length ratio, as provided by IMGT/V-QUEST.
For pseudogenes, the above information obtained using the option 'Search for insertions and deletions in V-REGION', with summary of the out-of-frame defects (for example, '1 del (1), 2 ins (1,2)' for one deletion of 1 nucleotide (nt), 2 insertions of 1 nt and 2nt).
IG and TR subgroup numbers
The IG and TR subgroup numbers are assigned, whenever it is possible, by comparison to the Homo sapiens subgroups (V-REGION nucleotide sequence identity >75% for functional and ORF genes and, for information, CDR-IMGT lengths). This means that some Homo sapiens subgroup numbers may not be represented in some species or conversely that new numbers may be added for subgroups not represented in Homo sapiens.
IG and TR gene numbers.
More information
Lefranc M-P. Immunoglobulin (IG) and T cell receptor genes (TR): IMGT® and the birth and rise of immunoinformatics. Front Immunol. 2014 Feb 05;5:22. doi: 10.3389/fimmu.2014.00022. Open access. PMID:24600447